| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
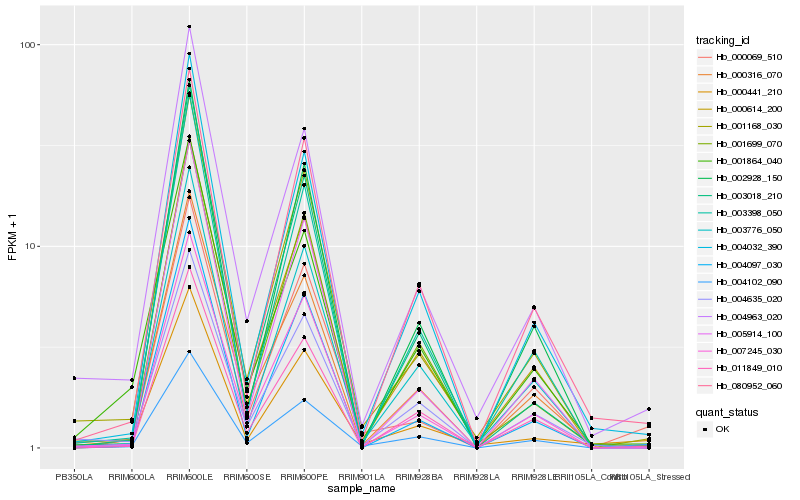
Hb_001168_030 |
0.0 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 2 |
Hb_007245_030 |
0.0662533173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004963_020 |
0.0700775785 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 4 |
Hb_000316_070 |
0.076599831 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 5 |
Hb_005914_100 |
0.0769519683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000614_200 |
0.0784177345 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 7 |
Hb_003776_050 |
0.0793295077 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 8 |
Hb_001699_070 |
0.080799483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003398_050 |
0.0840400674 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 10 |
Hb_004097_030 |
0.0879454149 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 11 |
Hb_002928_150 |
0.0891630551 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 12 |
Hb_004032_390 |
0.0902702914 |
- |
- |
CDK, putative [Ricinus communis] |
| 13 |
Hb_004635_020 |
0.0905159323 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 14 |
Hb_080952_060 |
0.0929992036 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 15 |
Hb_000069_510 |
0.0941592177 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 16 |
Hb_001864_040 |
0.0979626152 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 17 |
Hb_011849_010 |
0.098686353 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 18 |
Hb_000441_210 |
0.0990493642 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 19 |
Hb_004102_090 |
0.0990539346 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 20 |
Hb_003018_210 |
0.0991608951 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |