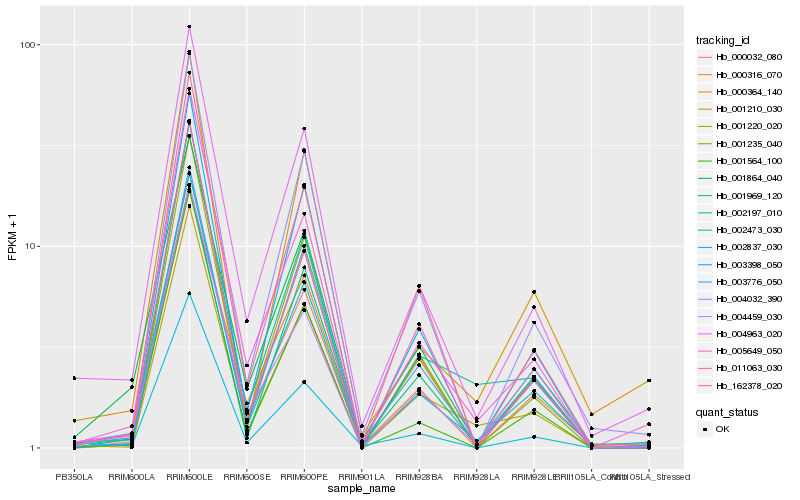
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002473_030 |
0.0 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 2 |
Hb_005649_050 |
0.0585213422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004963_020 |
0.064966248 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 4 |
Hb_004032_390 |
0.0722782404 |
- |
- |
CDK, putative [Ricinus communis] |
| 5 |
Hb_001220_020 |
0.079849735 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 6 |
Hb_002837_030 |
0.0810769626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003398_050 |
0.0817955558 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 8 |
Hb_162378_020 |
0.083039645 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 9 |
Hb_001564_100 |
0.0850016485 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 10 |
Hb_004459_030 |
0.0859748354 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000364_140 |
0.0859956169 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 12 |
Hb_001210_030 |
0.0867049052 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 13 |
Hb_001864_040 |
0.0876521984 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 14 |
Hb_000316_070 |
0.0886371035 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 15 |
Hb_002197_010 |
0.0888750131 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 16 |
Hb_001969_120 |
0.092908132 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 17 |
Hb_003776_050 |
0.0935646123 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 18 |
Hb_011063_030 |
0.0957518433 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_001235_040 |
0.0967679902 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 20 |
Hb_000032_080 |
0.0968336999 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |