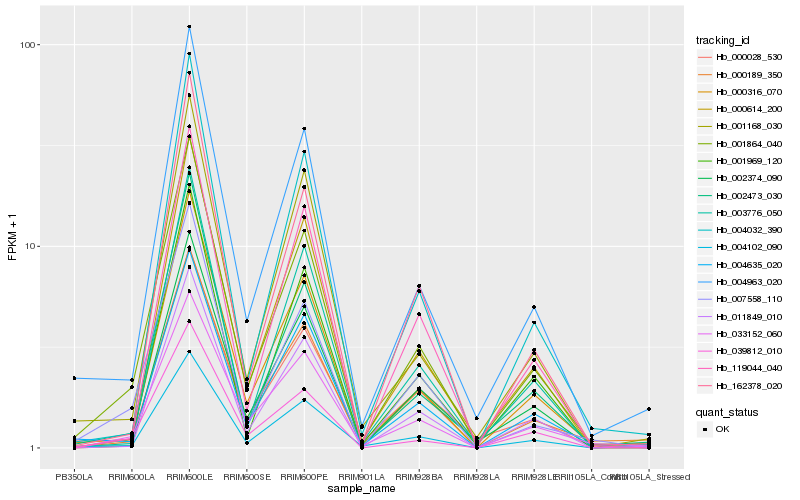
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001864_040 |
0.0 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 2 |
Hb_004963_020 |
0.0713941535 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 3 |
Hb_000316_070 |
0.073478667 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 4 |
Hb_007558_110 |
0.0821107309 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 5 |
Hb_002473_030 |
0.0876521984 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 6 |
Hb_004635_020 |
0.0879419783 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 7 |
Hb_033152_060 |
0.0933407272 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 8 |
Hb_004102_090 |
0.0972010024 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 9 |
Hb_039812_010 |
0.0972400307 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_001168_030 |
0.0979626152 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 11 |
Hb_000028_530 |
0.0986888864 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 12 |
Hb_004032_390 |
0.0990492241 |
- |
- |
CDK, putative [Ricinus communis] |
| 13 |
Hb_011849_010 |
0.0993311787 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 14 |
Hb_000189_350 |
0.1020915289 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 15 |
Hb_002374_090 |
0.1024387438 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 16 |
Hb_003776_050 |
0.1037106197 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 17 |
Hb_162378_020 |
0.1053346151 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 18 |
Hb_119044_040 |
0.1055905579 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 19 |
Hb_001969_120 |
0.1057282705 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 20 |
Hb_000614_200 |
0.1063018427 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |