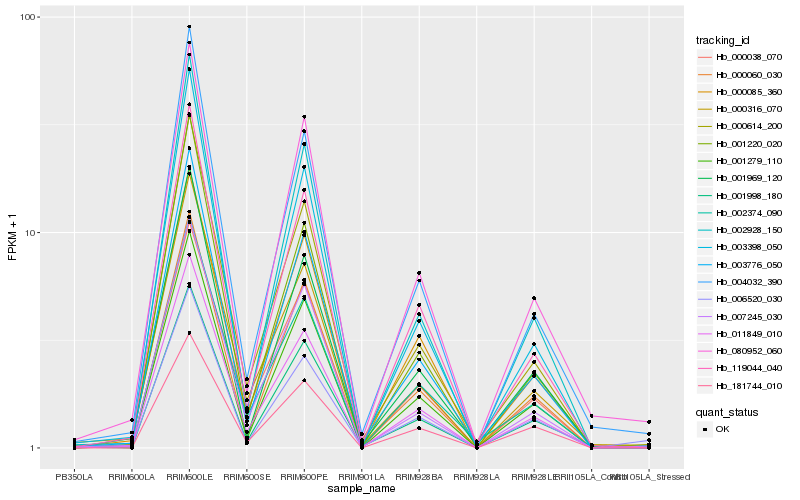
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011849_010 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000614_200 |
0.0475162861 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 3 |
Hb_003776_050 |
0.0558704328 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 4 |
Hb_002374_090 |
0.0581806866 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 5 |
Hb_000038_070 |
0.0591354999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_119044_040 |
0.05967643 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 7 |
Hb_000316_070 |
0.0606518834 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 8 |
Hb_001279_110 |
0.0630944747 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 9 |
Hb_000085_360 |
0.067504127 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 10 |
Hb_004032_390 |
0.0688722156 |
- |
- |
CDK, putative [Ricinus communis] |
| 11 |
Hb_007245_030 |
0.0728907503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002928_150 |
0.0747557326 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 13 |
Hb_001998_180 |
0.0747828075 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 14 |
Hb_000060_030 |
0.0754981868 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like [Jatropha curcas] |
| 15 |
Hb_080952_060 |
0.0756817082 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 16 |
Hb_001969_120 |
0.0757496545 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 17 |
Hb_003398_050 |
0.0762970112 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 18 |
Hb_001220_020 |
0.076993392 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 19 |
Hb_006520_030 |
0.0776794381 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 20 |
Hb_181744_010 |
0.078648687 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |