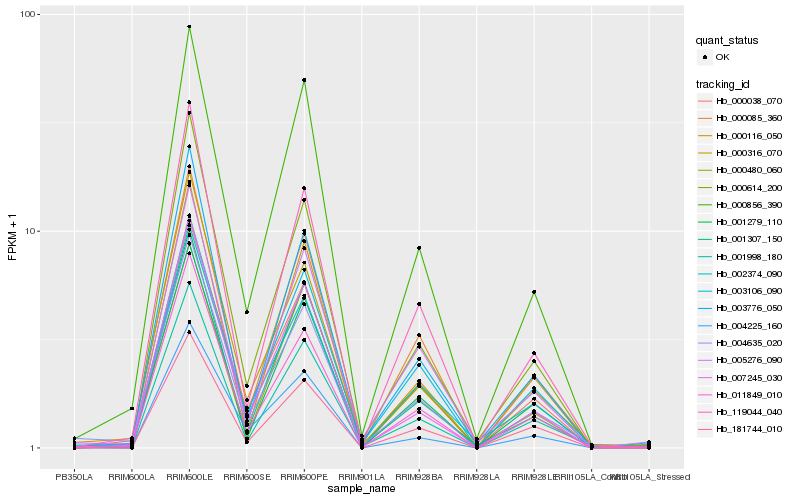
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000038_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011849_010 |
0.0591354999 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 3 |
Hb_000085_360 |
0.063586301 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 4 |
Hb_000856_390 |
0.0660920801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002374_090 |
0.0725541835 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 6 |
Hb_181744_010 |
0.0741896313 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 7 |
Hb_003106_090 |
0.0780285113 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 8 |
Hb_000614_200 |
0.0799401676 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 9 |
Hb_001279_110 |
0.0802857277 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 10 |
Hb_007245_030 |
0.0824469147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_119044_040 |
0.0867315382 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 12 |
Hb_000316_070 |
0.0875065396 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 13 |
Hb_001307_150 |
0.08759986 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_004225_160 |
0.0881167605 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 15 |
Hb_001998_180 |
0.0906020456 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 16 |
Hb_004635_020 |
0.0915333775 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 17 |
Hb_005276_090 |
0.0915494067 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000116_050 |
0.0936142228 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000480_060 |
0.0937883786 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 20 |
Hb_003776_050 |
0.0938973309 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |