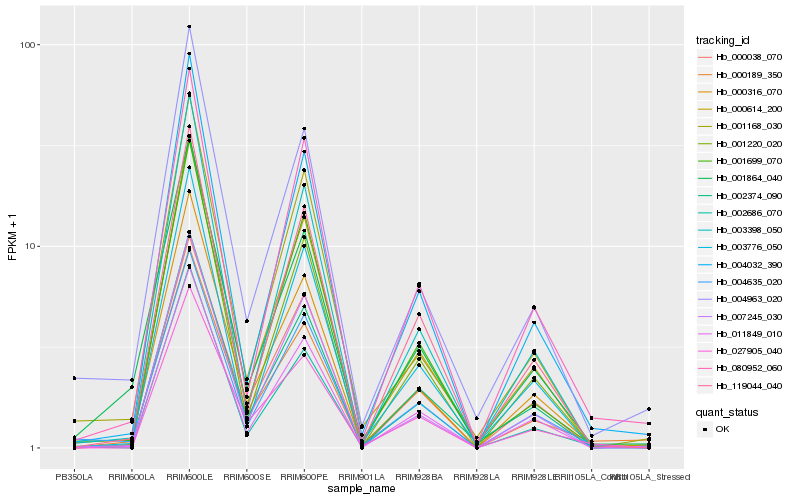
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000316_070 |
0.0 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 2 |
Hb_011849_010 |
0.0606518834 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 3 |
Hb_004635_020 |
0.0634504169 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 4 |
Hb_004963_020 |
0.0655756785 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 5 |
Hb_000614_200 |
0.0666719618 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 6 |
Hb_004032_390 |
0.0676451125 |
- |
- |
CDK, putative [Ricinus communis] |
| 7 |
Hb_007245_030 |
0.0685129737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001864_040 |
0.073478667 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 9 |
Hb_002374_090 |
0.0750387055 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 10 |
Hb_001168_030 |
0.076599831 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 11 |
Hb_003776_050 |
0.0800871869 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 12 |
Hb_000189_350 |
0.0844259057 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 13 |
Hb_003398_050 |
0.0854122304 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 14 |
Hb_027905_040 |
0.0861548766 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 15 |
Hb_001220_020 |
0.087237008 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 16 |
Hb_119044_040 |
0.0872915958 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 17 |
Hb_080952_060 |
0.0874715874 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 18 |
Hb_000038_070 |
0.0875065396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002686_070 |
0.0875652753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001699_070 |
0.0876912261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |