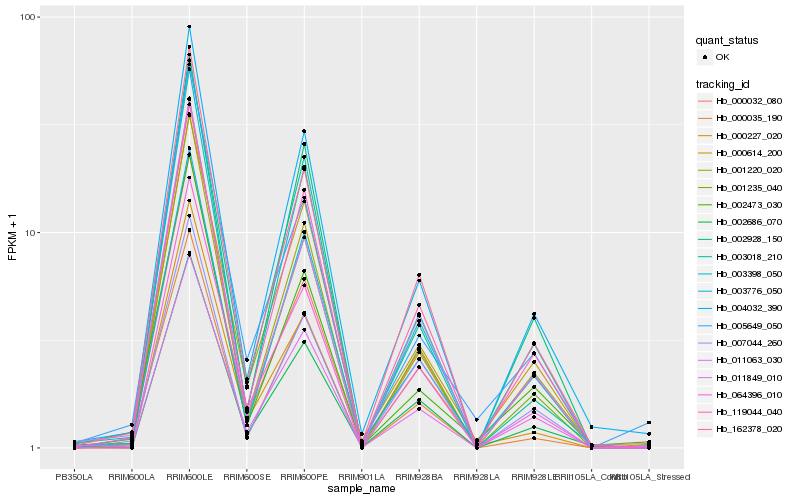
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162378_020 |
0.0 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 2 |
Hb_001220_020 |
0.0501053323 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 3 |
Hb_011063_030 |
0.0502820851 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000032_080 |
0.0553185763 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_004032_390 |
0.0555741105 |
- |
- |
CDK, putative [Ricinus communis] |
| 6 |
Hb_064396_010 |
0.056471392 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_002686_070 |
0.0592473197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001235_040 |
0.0645596553 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 9 |
Hb_003398_050 |
0.0695418191 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 10 |
Hb_119044_040 |
0.0768208056 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 11 |
Hb_000227_020 |
0.0782126023 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 12 |
Hb_011849_010 |
0.0817907911 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 13 |
Hb_003776_050 |
0.0828059885 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 14 |
Hb_002473_030 |
0.083039645 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 15 |
Hb_007044_260 |
0.0840090666 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000035_190 |
0.0861750426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003018_210 |
0.0871710816 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 18 |
Hb_002928_150 |
0.0889533709 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 19 |
Hb_000614_200 |
0.0890124343 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 20 |
Hb_005649_050 |
0.0897235697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |