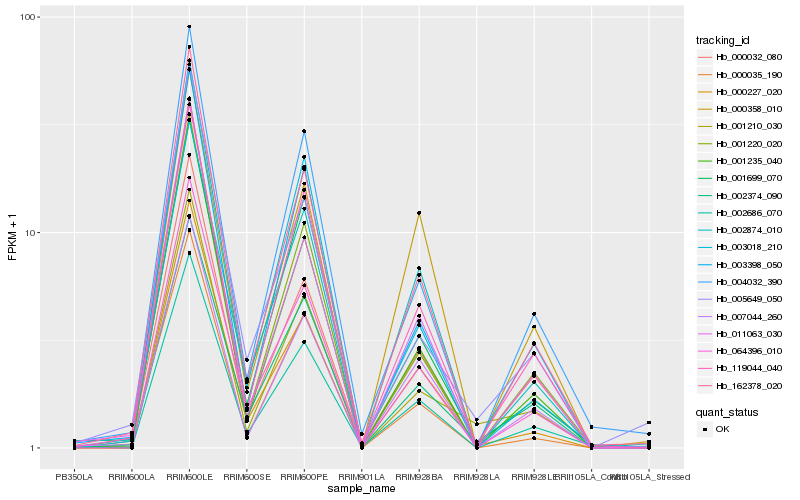
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_020 |
0.0 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 2 |
Hb_064396_010 |
0.057701167 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_162378_020 |
0.0782126023 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 4 |
Hb_002686_070 |
0.082922374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011063_030 |
0.0881632413 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_001235_040 |
0.0974813391 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 7 |
Hb_000032_080 |
0.1000059323 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002874_010 |
0.1062509558 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 9 |
Hb_007044_260 |
0.1063101165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001220_020 |
0.1075613184 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 11 |
Hb_000035_190 |
0.1133383299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001210_030 |
0.1139358628 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 13 |
Hb_000358_010 |
0.1147843519 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 14 |
Hb_119044_040 |
0.1151563628 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 15 |
Hb_003018_210 |
0.1177120536 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 16 |
Hb_004032_390 |
0.1192582533 |
- |
- |
CDK, putative [Ricinus communis] |
| 17 |
Hb_005649_050 |
0.1201580452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002374_090 |
0.1207205809 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 19 |
Hb_001699_070 |
0.1221190926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003398_050 |
0.1223203531 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |