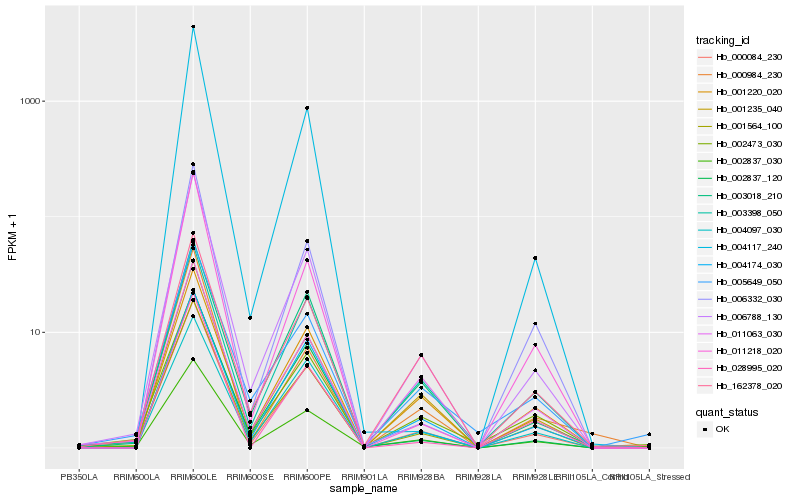
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_130 |
0.0 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 2 |
Hb_000084_230 |
0.0265841638 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 3 |
Hb_001564_100 |
0.0398587935 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 4 |
Hb_001235_040 |
0.052062743 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 5 |
Hb_001220_020 |
0.0863130019 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 6 |
Hb_004174_030 |
0.0899939223 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Populus euphratica] |
| 7 |
Hb_003018_210 |
0.0941769641 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 8 |
Hb_002837_120 |
0.0964679249 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 9 |
Hb_011218_020 |
0.0965517318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_028995_020 |
0.0969777064 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 11 |
Hb_005649_050 |
0.0989789638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006332_030 |
0.099420191 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 13 |
Hb_011063_030 |
0.0997190362 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_162378_020 |
0.1005319256 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 15 |
Hb_002473_030 |
0.1010033302 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 16 |
Hb_004097_030 |
0.1017747466 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 17 |
Hb_003398_050 |
0.1024112015 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 18 |
Hb_000984_230 |
0.1032075971 |
- |
- |
- |
| 19 |
Hb_004117_240 |
0.1041154041 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |
| 20 |
Hb_002837_030 |
0.1048853769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |