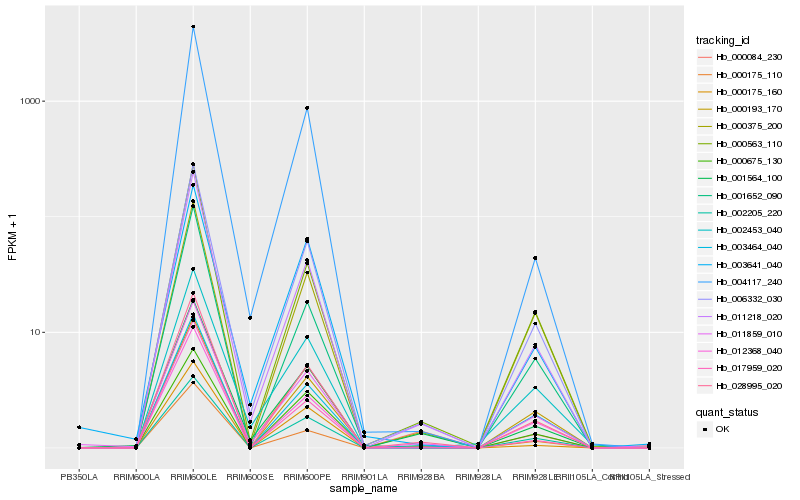
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006332_030 |
0.0 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 2 |
Hb_011218_020 |
0.04161749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_028995_020 |
0.0453746033 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 4 |
Hb_011859_010 |
0.0639254668 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 5 |
Hb_012368_040 |
0.0718699091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003464_040 |
0.0725562236 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 7 |
Hb_000563_110 |
0.0755613291 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 8 |
Hb_001652_090 |
0.0775195982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004117_240 |
0.0776223417 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |
| 10 |
Hb_000175_110 |
0.0792108705 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 11 |
Hb_001564_100 |
0.0806674622 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 12 |
Hb_000193_170 |
0.0816592568 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 13 |
Hb_002453_040 |
0.0829610591 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 14 |
Hb_002205_220 |
0.0862927667 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 15 |
Hb_000084_230 |
0.0912995428 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 16 |
Hb_000675_130 |
0.0915379881 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000375_200 |
0.0928196408 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 18 |
Hb_000175_160 |
0.093883893 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 19 |
Hb_003641_040 |
0.0942661317 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 20 |
Hb_017959_020 |
0.0946247486 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |