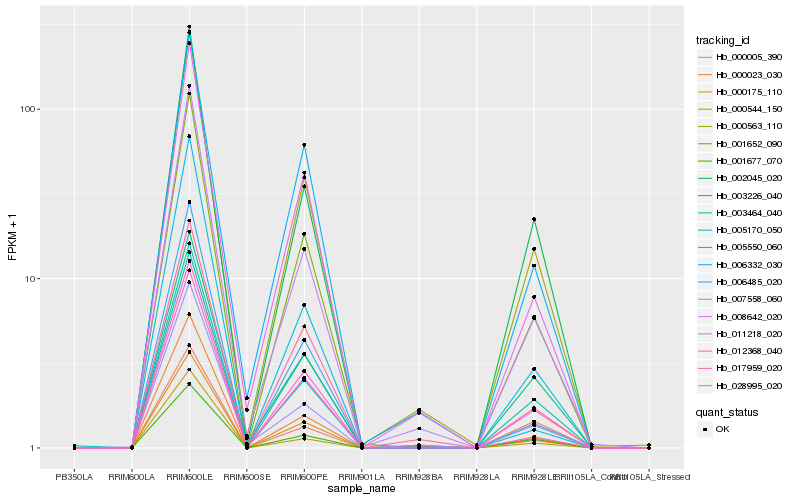
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 2 |
Hb_001652_090 |
0.0417445351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008642_020 |
0.0480489229 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 4 |
Hb_000175_110 |
0.0502864607 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 5 |
Hb_000005_390 |
0.0532577069 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 6 |
Hb_002045_020 |
0.0552257924 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 7 |
Hb_012368_040 |
0.0596170592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011218_020 |
0.0599499534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007558_060 |
0.0624313931 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 10 |
Hb_003464_040 |
0.0678025247 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 11 |
Hb_028995_020 |
0.0709543754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 12 |
Hb_003226_040 |
0.0723983598 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 13 |
Hb_000023_030 |
0.0725477946 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 14 |
Hb_006332_030 |
0.0755613291 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 15 |
Hb_005550_060 |
0.0772051047 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_005170_050 |
0.0831543809 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 17 |
Hb_017959_020 |
0.0833852328 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 18 |
Hb_001677_070 |
0.0869317036 |
- |
- |
PREDICTED: putative caffeoyl-CoA O-methyltransferase At1g67980 [Jatropha curcas] |
| 19 |
Hb_006485_020 |
0.0882933642 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 20 |
Hb_000544_150 |
0.0891097636 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |