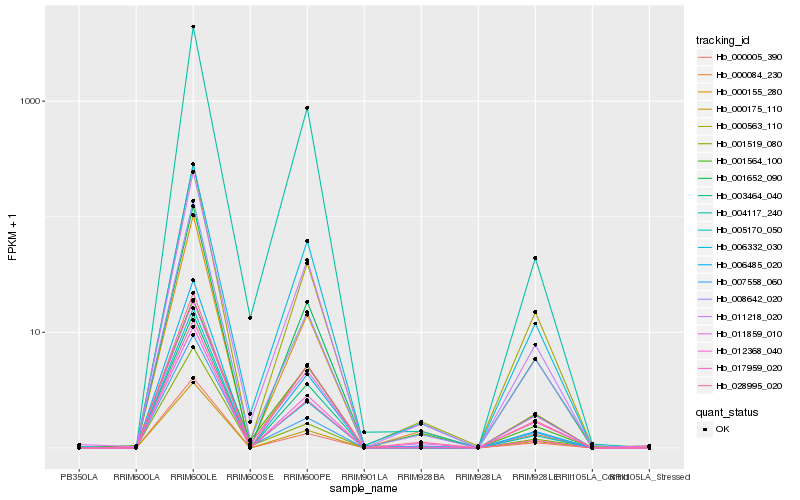
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011218_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006332_030 |
0.04161749 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 3 |
Hb_028995_020 |
0.0486609832 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 4 |
Hb_017959_020 |
0.0594697201 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 5 |
Hb_000563_110 |
0.0599499534 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 6 |
Hb_001652_090 |
0.0613478612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004117_240 |
0.0657944413 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |
| 8 |
Hb_000175_110 |
0.0724477674 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 9 |
Hb_008642_020 |
0.0726131587 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 10 |
Hb_006485_020 |
0.0737652326 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 11 |
Hb_000155_280 |
0.0772101188 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 12 |
Hb_000005_390 |
0.077576014 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 13 |
Hb_011859_010 |
0.0782616109 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 14 |
Hb_005170_050 |
0.0800867725 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 15 |
Hb_012368_040 |
0.0820816102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007558_060 |
0.0838318102 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 17 |
Hb_003464_040 |
0.0874988656 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_001564_100 |
0.0891714511 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 19 |
Hb_000084_230 |
0.09271833 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 20 |
Hb_001519_080 |
0.0956612765 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |