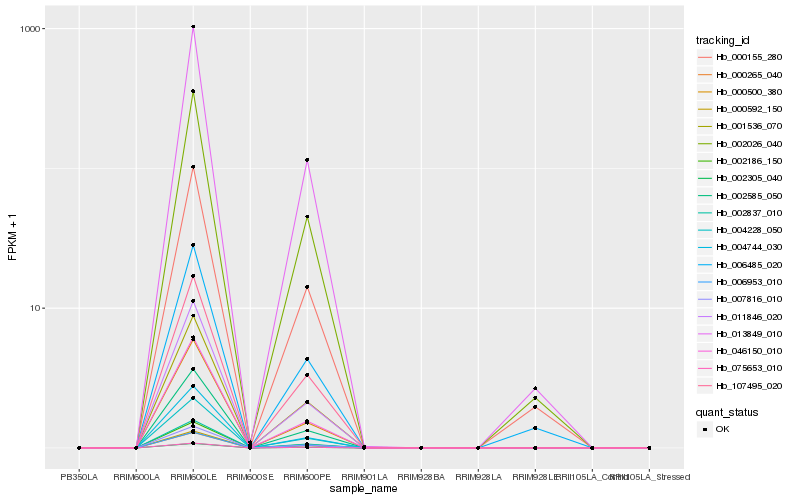
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002026_040 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 2 |
Hb_013849_010 |
0.0229643325 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 3 |
Hb_000155_280 |
0.0302992498 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 4 |
Hb_002585_050 |
0.0416941405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007816_010 |
0.0422718009 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 6 |
Hb_000592_150 |
0.042812751 |
- |
- |
Os05g0413200 [Oryza sativa Japonica Group] |
| 7 |
Hb_004228_050 |
0.0430348792 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 8 |
Hb_002837_010 |
0.0433587696 |
- |
- |
hypothetical protein POPTR_0497s00200g [Populus trichocarpa] |
| 9 |
Hb_002305_040 |
0.0447540884 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 10 |
Hb_075653_010 |
0.0451172921 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 11 |
Hb_011846_020 |
0.045221645 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 12 |
Hb_002186_150 |
0.0459511035 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 13 |
Hb_046150_010 |
0.0459651949 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 6-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_000265_040 |
0.0467963402 |
- |
- |
PREDICTED: subtilisin-like protease, partial [Prunus mume] |
| 15 |
Hb_001536_070 |
0.0468487565 |
- |
- |
hypothetical protein JCGZ_20053 [Jatropha curcas] |
| 16 |
Hb_107495_020 |
0.0471919308 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 17 |
Hb_004744_030 |
0.0472445034 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 18 |
Hb_000500_380 |
0.0475629477 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45670-like [Jatropha curcas] |
| 19 |
Hb_006485_020 |
0.0479269133 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 20 |
Hb_006953_010 |
0.0485675992 |
- |
- |
cytochrome P450, putative [Ricinus communis] |