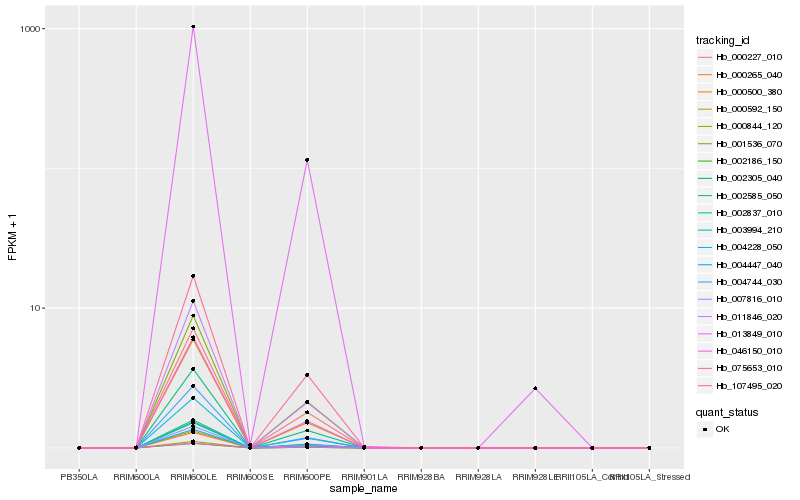
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007816_010 |
0.0 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 2 |
Hb_002837_010 |
0.0049234411 |
- |
- |
hypothetical protein POPTR_0497s00200g [Populus trichocarpa] |
| 3 |
Hb_002585_050 |
0.0077373317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011846_020 |
0.0105334549 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 5 |
Hb_002186_150 |
0.0123401168 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 6 |
Hb_046150_010 |
0.0123736312 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 6-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_004744_030 |
0.0152440171 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_000500_380 |
0.0159138424 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45670-like [Jatropha curcas] |
| 9 |
Hb_000592_150 |
0.0167653514 |
- |
- |
Os05g0413200 [Oryza sativa Japonica Group] |
| 10 |
Hb_004228_050 |
0.0176991816 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 11 |
Hb_002305_040 |
0.0232997806 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 12 |
Hb_075653_010 |
0.0242736528 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 13 |
Hb_004447_040 |
0.0263967825 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 14 |
Hb_003994_210 |
0.0278816907 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000227_010 |
0.0282211643 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 16 |
Hb_000265_040 |
0.0282825062 |
- |
- |
PREDICTED: subtilisin-like protease, partial [Prunus mume] |
| 17 |
Hb_001536_070 |
0.0283976809 |
- |
- |
hypothetical protein JCGZ_20053 [Jatropha curcas] |
| 18 |
Hb_000844_120 |
0.0284644274 |
- |
- |
hypothetical protein JCGZ_09179 [Jatropha curcas] |
| 19 |
Hb_013849_010 |
0.0288355855 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 20 |
Hb_107495_020 |
0.0291399975 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |