| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
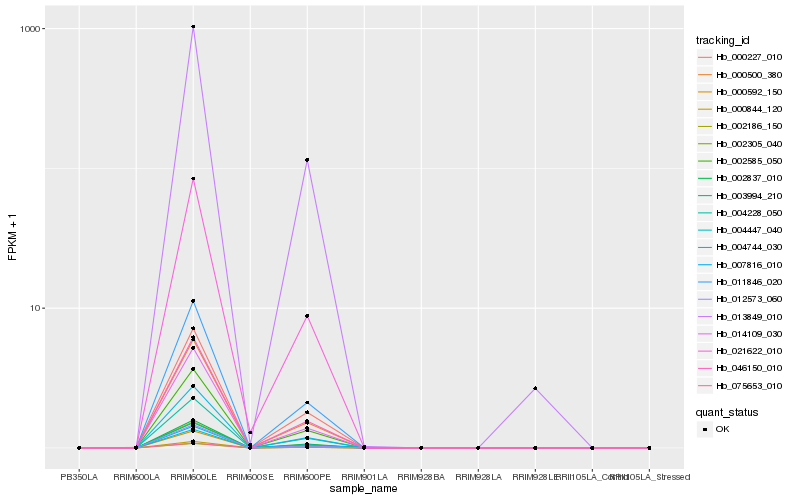
Hb_000500_380 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45670-like [Jatropha curcas] |
| 2 |
Hb_004744_030 |
0.0006700878 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 3 |
Hb_046150_010 |
0.0035413287 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 6-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_002186_150 |
0.0035748506 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 5 |
Hb_011846_020 |
0.0053818264 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 6 |
Hb_004447_040 |
0.0104902624 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 7 |
Hb_002837_010 |
0.0109917554 |
- |
- |
hypothetical protein POPTR_0497s00200g [Populus trichocarpa] |
| 8 |
Hb_003994_210 |
0.0119767126 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000844_120 |
0.0125600897 |
- |
- |
hypothetical protein JCGZ_09179 [Jatropha curcas] |
| 10 |
Hb_014109_030 |
0.0152694124 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 11 |
Hb_007816_010 |
0.0159138424 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 12 |
Hb_002585_050 |
0.023646741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_013849_010 |
0.0286386726 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 14 |
Hb_000592_150 |
0.0326662218 |
- |
- |
Os05g0413200 [Oryza sativa Japonica Group] |
| 15 |
Hb_004228_050 |
0.0335989713 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 16 |
Hb_000227_010 |
0.0378930541 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 17 |
Hb_002305_040 |
0.0391923434 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 18 |
Hb_075653_010 |
0.0401648311 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 19 |
Hb_021622_010 |
0.0418712885 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 20 |
Hb_012573_060 |
0.043943967 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |