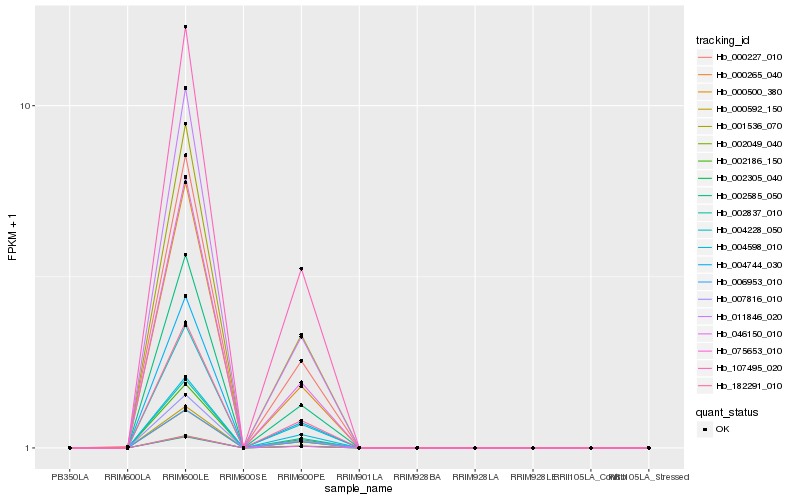
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002585_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007816_010 |
0.0077373317 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 3 |
Hb_000592_150 |
0.0090296653 |
- |
- |
Os05g0413200 [Oryza sativa Japonica Group] |
| 4 |
Hb_004228_050 |
0.0099637625 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 5 |
Hb_002837_010 |
0.0126600583 |
- |
- |
hypothetical protein POPTR_0497s00200g [Populus trichocarpa] |
| 6 |
Hb_002305_040 |
0.0155663296 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 7 |
Hb_075653_010 |
0.0165406069 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 8 |
Hb_011846_020 |
0.0182685498 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 9 |
Hb_002186_150 |
0.0200745561 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 10 |
Hb_046150_010 |
0.0201080575 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 6-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_000265_040 |
0.0205513193 |
- |
- |
PREDICTED: subtilisin-like protease, partial [Prunus mume] |
| 12 |
Hb_001536_070 |
0.0206665519 |
- |
- |
hypothetical protein JCGZ_20053 [Jatropha curcas] |
| 13 |
Hb_107495_020 |
0.0214092476 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 14 |
Hb_004744_030 |
0.0229772293 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 15 |
Hb_000500_380 |
0.023646741 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45670-like [Jatropha curcas] |
| 16 |
Hb_006953_010 |
0.024212561 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_000227_010 |
0.0258495411 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 18 |
Hb_002049_040 |
0.0262934273 |
- |
- |
PREDICTED: protein UNUSUAL FLORAL ORGANS [Jatropha curcas] |
| 19 |
Hb_182291_010 |
0.027364725 |
- |
- |
hypothetical protein CICLE_v10029654mg [Citrus clementina] |
| 20 |
Hb_004598_010 |
0.0285871865 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |