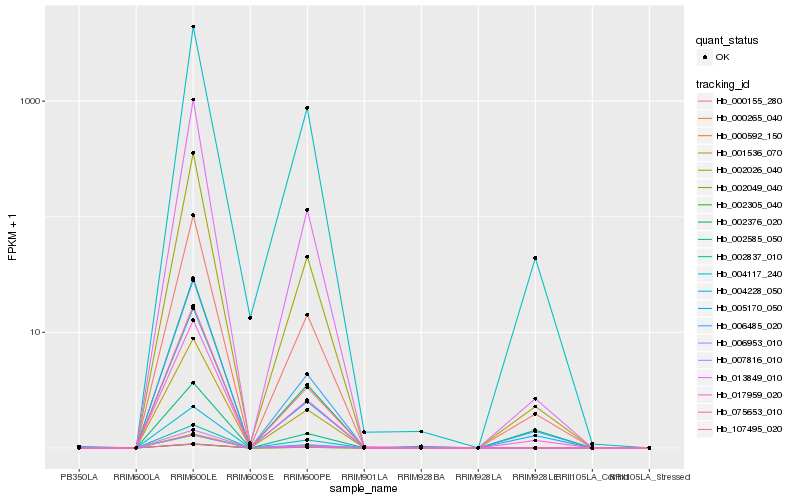
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000155_280 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 2 |
Hb_006485_020 |
0.0211524632 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 3 |
Hb_002026_040 |
0.0302992498 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 4 |
Hb_017959_020 |
0.0307531932 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 5 |
Hb_013849_010 |
0.0491113354 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 6 |
Hb_002376_020 |
0.0527848271 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 7 |
Hb_005170_050 |
0.0642429385 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 8 |
Hb_000592_150 |
0.0653579213 |
- |
- |
Os05g0413200 [Oryza sativa Japonica Group] |
| 9 |
Hb_004228_050 |
0.0654210287 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 10 |
Hb_002585_050 |
0.065432964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002305_040 |
0.0660748605 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 12 |
Hb_075653_010 |
0.0662360984 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 13 |
Hb_007816_010 |
0.0664759585 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 14 |
Hb_000265_040 |
0.0670444539 |
- |
- |
PREDICTED: subtilisin-like protease, partial [Prunus mume] |
| 15 |
Hb_001536_070 |
0.0670710658 |
- |
- |
hypothetical protein JCGZ_20053 [Jatropha curcas] |
| 16 |
Hb_107495_020 |
0.0672470522 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 17 |
Hb_002837_010 |
0.0675915962 |
- |
- |
hypothetical protein POPTR_0497s00200g [Populus trichocarpa] |
| 18 |
Hb_006953_010 |
0.0679800851 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_002049_040 |
0.0685929867 |
- |
- |
PREDICTED: protein UNUSUAL FLORAL ORGANS [Jatropha curcas] |
| 20 |
Hb_004117_240 |
0.068859884 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |