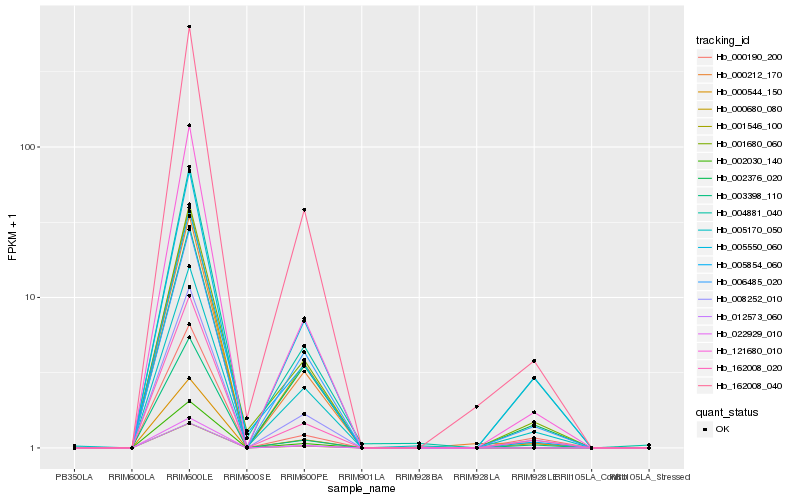
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008252_010 |
0.0 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 2 |
Hb_162008_020 |
0.0239665855 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 3 |
Hb_002376_020 |
0.0371547795 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 4 |
Hb_121680_010 |
0.0438150336 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 5 |
Hb_162008_040 |
0.048046622 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 6 |
Hb_005550_060 |
0.05776755 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000212_170 |
0.0580538233 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 8 |
Hb_001680_060 |
0.0591283288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000544_150 |
0.0645047295 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 10 |
Hb_004881_040 |
0.0668045949 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 11 |
Hb_005170_050 |
0.0720884653 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 12 |
Hb_005854_060 |
0.0736380912 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_000190_200 |
0.0736493559 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 14 |
Hb_001546_100 |
0.0745400703 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 15 |
Hb_002030_140 |
0.074731942 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 16 |
Hb_022929_010 |
0.0748500572 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 17 |
Hb_000680_080 |
0.0748964758 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 18 |
Hb_012573_060 |
0.075380289 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |
| 19 |
Hb_006485_020 |
0.0754516985 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 20 |
Hb_003398_110 |
0.0754929713 |
- |
- |
PREDICTED: uncharacterized protein LOC105647583 [Jatropha curcas] |