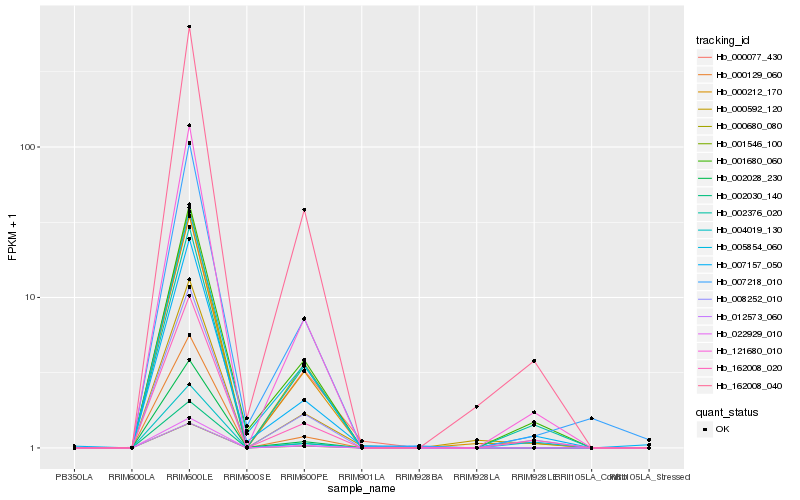
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162008_040 |
0.0 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 2 |
Hb_000212_170 |
0.0285297858 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 3 |
Hb_121680_010 |
0.0400765063 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 4 |
Hb_008252_010 |
0.048046622 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 5 |
Hb_005854_060 |
0.0529431878 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_162008_020 |
0.0546297278 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 7 |
Hb_001546_100 |
0.0565887513 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 8 |
Hb_002030_140 |
0.0575586566 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 9 |
Hb_022929_010 |
0.0578815122 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 10 |
Hb_000680_080 |
0.0579989662 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 11 |
Hb_012573_060 |
0.0590739907 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |
| 12 |
Hb_004019_130 |
0.0627481718 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 13 |
Hb_001680_060 |
0.0633622393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000129_060 |
0.0654791102 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000592_120 |
0.0655172265 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 16 |
Hb_000077_430 |
0.0693484186 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 17 |
Hb_002376_020 |
0.0693511516 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 18 |
Hb_007218_010 |
0.0710191587 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 19 |
Hb_007157_050 |
0.0716797062 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 20 |
Hb_002028_230 |
0.0717059677 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |