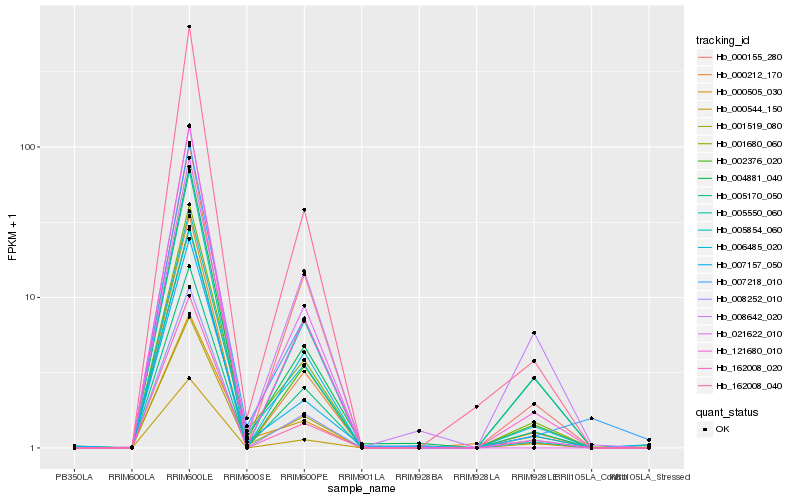
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001680_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005854_060 |
0.0469143678 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_008252_010 |
0.0591283288 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 4 |
Hb_162008_040 |
0.0633622393 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 5 |
Hb_002376_020 |
0.0644015734 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 6 |
Hb_162008_020 |
0.0673234693 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 7 |
Hb_007157_050 |
0.0679286189 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 8 |
Hb_004881_040 |
0.0697257001 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 9 |
Hb_005170_050 |
0.0754585959 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 10 |
Hb_121680_010 |
0.0758733444 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005550_060 |
0.0781730333 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000505_030 |
0.0822192395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007218_010 |
0.0823423617 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 14 |
Hb_000212_170 |
0.0824119613 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 15 |
Hb_001519_080 |
0.0836600814 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 16 |
Hb_021622_010 |
0.0839903574 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 17 |
Hb_000544_150 |
0.0860094209 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 18 |
Hb_006485_020 |
0.0875485189 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 19 |
Hb_008642_020 |
0.0907322089 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 20 |
Hb_000155_280 |
0.0918812284 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |