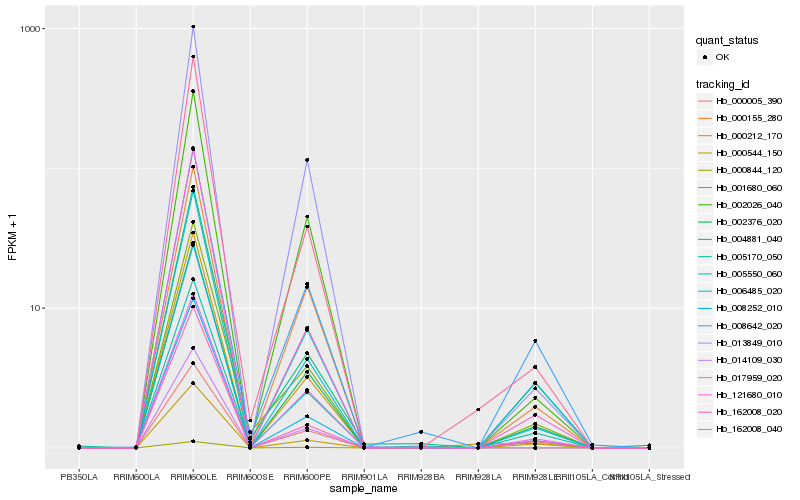
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002376_020 |
0.0 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 2 |
Hb_005550_060 |
0.0366387844 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_008252_010 |
0.0371547795 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 4 |
Hb_006485_020 |
0.0396811083 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 5 |
Hb_005170_050 |
0.0508741944 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 6 |
Hb_000155_280 |
0.0527848271 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 7 |
Hb_000005_390 |
0.0571799447 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 8 |
Hb_162008_020 |
0.0589829277 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 9 |
Hb_000544_150 |
0.059397876 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 10 |
Hb_017959_020 |
0.0595911439 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 11 |
Hb_001680_060 |
0.0644015734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002026_040 |
0.0662344533 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 13 |
Hb_008642_020 |
0.0686603699 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 14 |
Hb_162008_040 |
0.0693511516 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 15 |
Hb_013849_010 |
0.069509814 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 16 |
Hb_000212_170 |
0.0732519067 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 17 |
Hb_121680_010 |
0.078005758 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004881_040 |
0.0799965726 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 19 |
Hb_014109_030 |
0.0826182918 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 20 |
Hb_000844_120 |
0.0827877406 |
- |
- |
hypothetical protein JCGZ_09179 [Jatropha curcas] |