| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
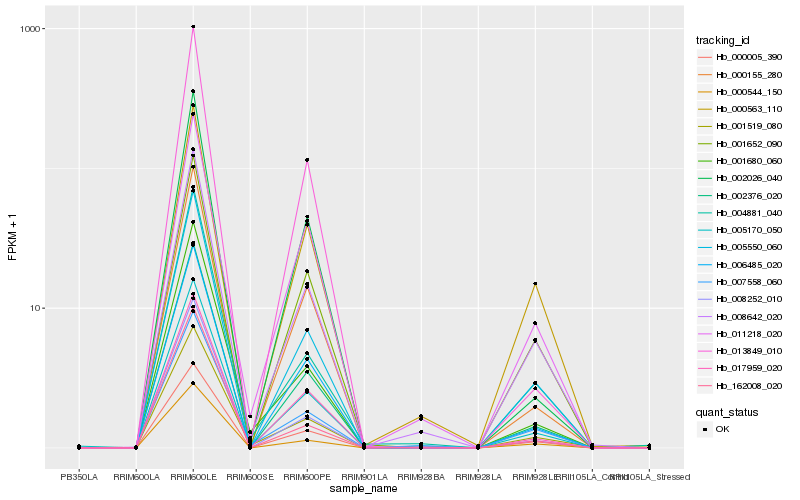
Hb_005170_050 |
0.0 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 2 |
Hb_002376_020 |
0.0508741944 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 3 |
Hb_008642_020 |
0.0531368884 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 4 |
Hb_006485_020 |
0.0556012136 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 5 |
Hb_005550_060 |
0.0565542009 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_017959_020 |
0.0566662837 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 7 |
Hb_000005_390 |
0.0637314476 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 8 |
Hb_000155_280 |
0.0642429385 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 9 |
Hb_001519_080 |
0.0696852912 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 10 |
Hb_007558_060 |
0.0715869047 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 11 |
Hb_008252_010 |
0.0720884653 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 12 |
Hb_001680_060 |
0.0754585959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000544_150 |
0.0754827986 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 14 |
Hb_002026_040 |
0.079425034 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 15 |
Hb_011218_020 |
0.0800867725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001652_090 |
0.0819994428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000563_110 |
0.0831543809 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 18 |
Hb_004881_040 |
0.0852568661 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 19 |
Hb_013849_010 |
0.0877156002 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 20 |
Hb_162008_020 |
0.0883789397 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |