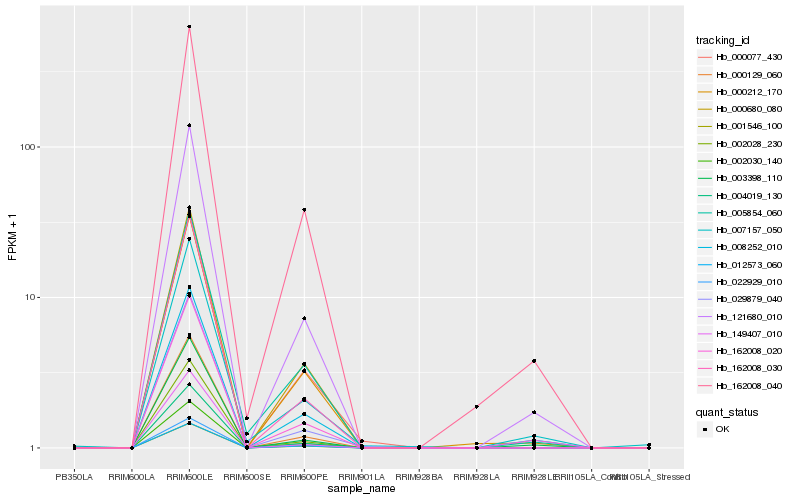
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121680_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 2 |
Hb_162008_020 |
0.0373253293 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 3 |
Hb_162008_040 |
0.0400765063 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 4 |
Hb_008252_010 |
0.0438150336 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 5 |
Hb_004019_130 |
0.0506931334 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 6 |
Hb_000129_060 |
0.0513536363 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000212_170 |
0.051818915 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 8 |
Hb_162008_030 |
0.0567459013 |
- |
- |
cytochrome p450 [Ageratina adenophora] |
| 9 |
Hb_003398_110 |
0.0571211379 |
- |
- |
PREDICTED: uncharacterized protein LOC105647583 [Jatropha curcas] |
| 10 |
Hb_001546_100 |
0.0592834536 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 11 |
Hb_002030_140 |
0.0626432071 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 12 |
Hb_022929_010 |
0.0635031949 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 13 |
Hb_000680_080 |
0.0638000294 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 14 |
Hb_002028_230 |
0.0645566846 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 15 |
Hb_012573_060 |
0.0662492742 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |
| 16 |
Hb_029879_040 |
0.0666231024 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 17 |
Hb_007157_050 |
0.067892956 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 18 |
Hb_005854_060 |
0.0708522315 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_149407_010 |
0.0711364774 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 20 |
Hb_000077_430 |
0.0733696302 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |