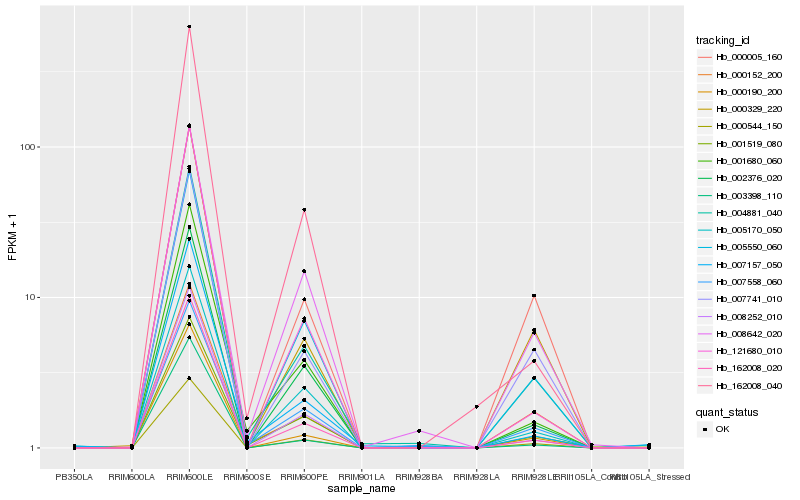
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004881_040 |
0.0 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 2 |
Hb_000190_200 |
0.0498533495 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 3 |
Hb_000544_150 |
0.0597464258 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 4 |
Hb_162008_020 |
0.0599794194 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 5 |
Hb_000329_220 |
0.0649690914 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 6 |
Hb_007157_050 |
0.0658833272 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 7 |
Hb_008252_010 |
0.0668045949 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 8 |
Hb_007741_010 |
0.0677431477 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 9 |
Hb_001680_060 |
0.0697257001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005550_060 |
0.0716853318 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_002376_020 |
0.0799965726 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 12 |
Hb_008642_020 |
0.0802735509 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 13 |
Hb_007558_060 |
0.0836292623 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 14 |
Hb_000152_200 |
0.0842294888 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_121680_010 |
0.0843120672 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005170_050 |
0.0852568661 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 17 |
Hb_001519_080 |
0.0880198319 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 18 |
Hb_003398_110 |
0.0889234727 |
- |
- |
PREDICTED: uncharacterized protein LOC105647583 [Jatropha curcas] |
| 19 |
Hb_162008_040 |
0.0892150624 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 20 |
Hb_000005_160 |
0.0919949203 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |