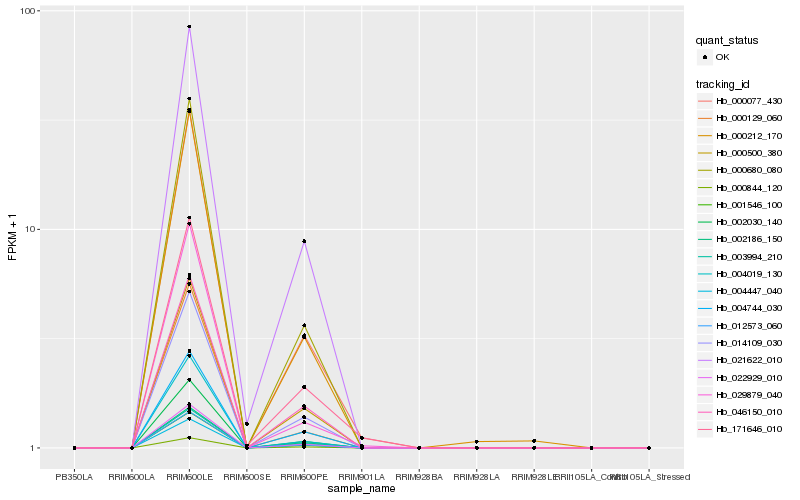
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_430 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 2 |
Hb_002030_140 |
0.0390072581 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 3 |
Hb_022929_010 |
0.0390619318 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 4 |
Hb_000680_080 |
0.0390926885 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 5 |
Hb_001546_100 |
0.0393606637 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 6 |
Hb_012573_060 |
0.0395568511 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |
| 7 |
Hb_014109_030 |
0.0525950933 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 8 |
Hb_000844_120 |
0.0544437344 |
- |
- |
hypothetical protein JCGZ_09179 [Jatropha curcas] |
| 9 |
Hb_003994_210 |
0.0548509037 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_004019_130 |
0.0552964487 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 11 |
Hb_004447_040 |
0.0559020862 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 12 |
Hb_171646_010 |
0.0591290799 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 13 |
Hb_000129_060 |
0.05947456 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000212_170 |
0.0595485334 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 15 |
Hb_029879_040 |
0.0631618504 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 16 |
Hb_000500_380 |
0.063798192 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45670-like [Jatropha curcas] |
| 17 |
Hb_004744_030 |
0.0643266936 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 18 |
Hb_046150_010 |
0.0666185353 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 6-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_002186_150 |
0.0666455424 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 20 |
Hb_021622_010 |
0.0672466261 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |