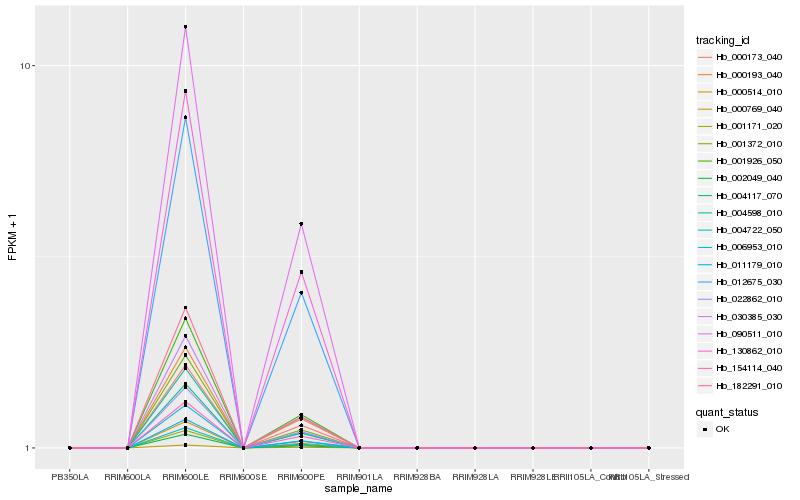
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_070 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 2 |
Hb_001926_050 |
0.008027121 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 3 |
Hb_030385_030 |
0.0122663426 |
- |
- |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011179_010 |
0.0156883308 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 5 |
Hb_004722_050 |
0.0179660435 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 6 |
Hb_000173_040 |
0.0217170615 |
- |
- |
putative xylanase inhibitor, partial [Triticum aestivum] |
| 7 |
Hb_154114_040 |
0.022423543 |
- |
- |
hypothetical protein B456_001G056600 [Gossypium raimondii] |
| 8 |
Hb_000193_040 |
0.0249968701 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 9 |
Hb_022862_010 |
0.0270307862 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 10 |
Hb_000514_010 |
0.0287570185 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 11 |
Hb_000769_040 |
0.0308218883 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 12 |
Hb_001171_020 |
0.0341623824 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 3-like [Citrus sinensis] |
| 13 |
Hb_012675_030 |
0.0356034899 |
- |
- |
hypothetical protein POPTR_0013s04780g [Populus trichocarpa] |
| 14 |
Hb_090511_010 |
0.0359103077 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001372_010 |
0.0371916342 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_004598_010 |
0.0375189764 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |
| 17 |
Hb_130862_010 |
0.0381587953 |
- |
- |
purine permease, putative [Ricinus communis] |
| 18 |
Hb_182291_010 |
0.0387407206 |
- |
- |
hypothetical protein CICLE_v10029654mg [Citrus clementina] |
| 19 |
Hb_002049_040 |
0.0398112013 |
- |
- |
PREDICTED: protein UNUSUAL FLORAL ORGANS [Jatropha curcas] |
| 20 |
Hb_006953_010 |
0.041889973 |
- |
- |
cytochrome P450, putative [Ricinus communis] |