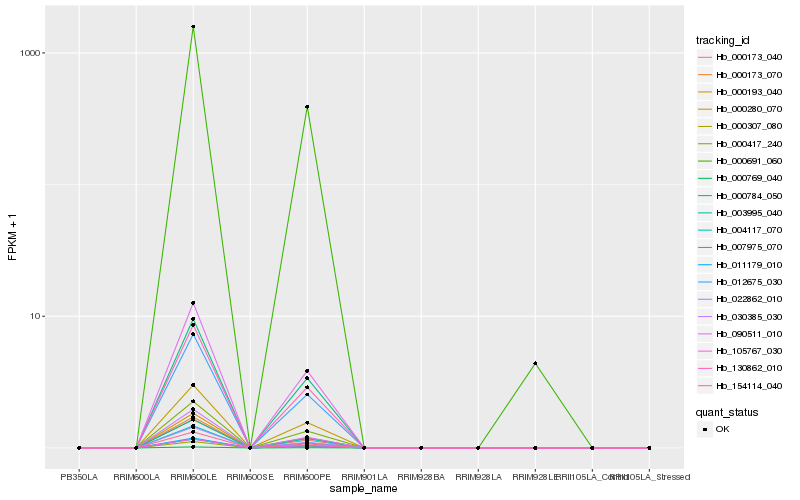
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022862_010 |
0.0 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 2 |
Hb_000193_040 |
0.0020350549 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 3 |
Hb_000769_040 |
0.0037936913 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 4 |
Hb_154114_040 |
0.0046095681 |
- |
- |
hypothetical protein B456_001G056600 [Gossypium raimondii] |
| 5 |
Hb_000173_040 |
0.005316325 |
- |
- |
putative xylanase inhibitor, partial [Triticum aestivum] |
| 6 |
Hb_012675_030 |
0.0085794082 |
- |
- |
hypothetical protein POPTR_0013s04780g [Populus trichocarpa] |
| 7 |
Hb_090511_010 |
0.0088865218 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_130862_010 |
0.0111372933 |
- |
- |
purine permease, putative [Ricinus communis] |
| 9 |
Hb_011179_010 |
0.01134651 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 10 |
Hb_030385_030 |
0.0147685965 |
- |
- |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 11 |
Hb_105767_030 |
0.0203950485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007975_070 |
0.0217989134 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 13 |
Hb_000417_240 |
0.0267424881 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 14 |
Hb_003995_040 |
0.0268825477 |
- |
- |
PREDICTED: tropinone reductase homolog At2g29290-like [Jatropha curcas] |
| 15 |
Hb_004117_070 |
0.0270307862 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 16 |
Hb_000173_070 |
0.0275626847 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 17 |
Hb_000307_080 |
0.0285539732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000280_070 |
0.0305146489 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_000691_060 |
0.0307532916 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 20 |
Hb_000784_050 |
0.0310390018 |
- |
- |
PREDICTED: auxin-induced protein X10A-like [Nelumbo nucifera] |