| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
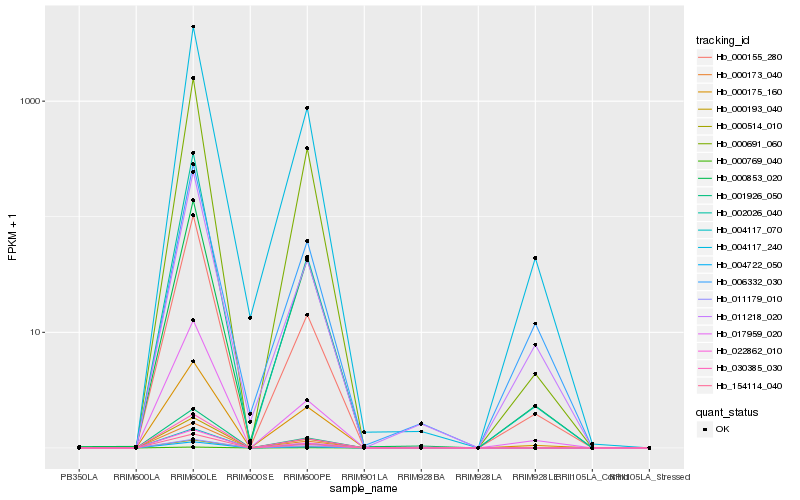
Hb_004117_240 |
0.0 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |
| 2 |
Hb_000691_060 |
0.0639729011 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 3 |
Hb_000175_160 |
0.0642224495 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 4 |
Hb_011218_020 |
0.0657944413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000155_280 |
0.068859884 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 6 |
Hb_017959_020 |
0.0703455536 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 7 |
Hb_004117_070 |
0.072593023 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 8 |
Hb_001926_050 |
0.0731623052 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 9 |
Hb_030385_030 |
0.0734204616 |
- |
- |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011179_010 |
0.0740112778 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 11 |
Hb_000853_020 |
0.0743775223 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 12 |
Hb_004722_050 |
0.0750539225 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 13 |
Hb_000173_040 |
0.0754174999 |
- |
- |
putative xylanase inhibitor, partial [Triticum aestivum] |
| 14 |
Hb_154114_040 |
0.0756118799 |
- |
- |
hypothetical protein B456_001G056600 [Gossypium raimondii] |
| 15 |
Hb_000193_040 |
0.0763706947 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 16 |
Hb_022862_010 |
0.0770256823 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 17 |
Hb_006332_030 |
0.0776223417 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 18 |
Hb_002026_040 |
0.078208496 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 19 |
Hb_000769_040 |
0.0783721605 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 20 |
Hb_000514_010 |
0.078485887 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |