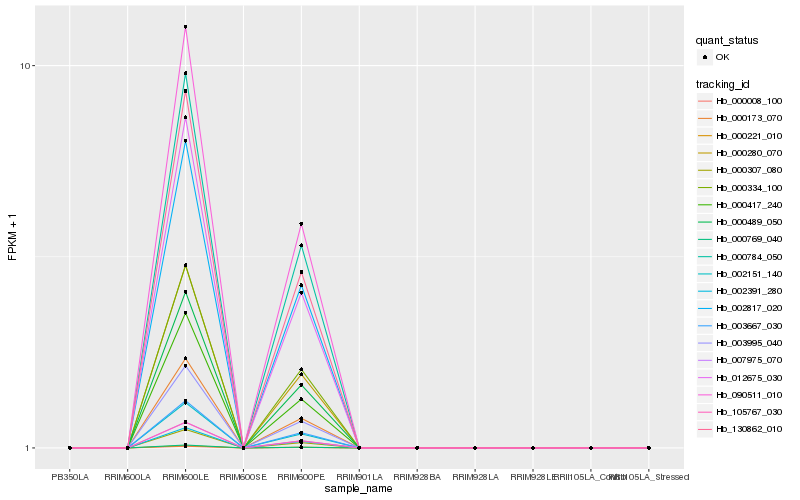
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_070 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 2 |
Hb_003995_040 |
0.000680497 |
- |
- |
PREDICTED: tropinone reductase homolog At2g29290-like [Jatropha curcas] |
| 3 |
Hb_000417_240 |
0.0008206285 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 4 |
Hb_000307_080 |
0.0009918442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000280_070 |
0.0029537269 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_000784_050 |
0.0034784268 |
- |
- |
PREDICTED: auxin-induced protein X10A-like [Nelumbo nucifera] |
| 7 |
Hb_002151_140 |
0.0052089493 |
- |
- |
PREDICTED: lichenase-2-like [Pyrus x bretschneideri] |
| 8 |
Hb_007975_070 |
0.0057662663 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 9 |
Hb_105767_030 |
0.007170546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003667_030 |
0.0119531713 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 11 |
Hb_000489_050 |
0.014044594 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 12 |
Hb_130862_010 |
0.0164290906 |
- |
- |
purine permease, putative [Ricinus communis] |
| 13 |
Hb_000334_100 |
0.0175546642 |
- |
- |
hypothetical protein JCGZ_16788 [Jatropha curcas] |
| 14 |
Hb_090511_010 |
0.0186795319 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_012675_030 |
0.0189865843 |
- |
- |
hypothetical protein POPTR_0013s04780g [Populus trichocarpa] |
| 16 |
Hb_000008_100 |
0.0211278636 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 17 |
Hb_002391_280 |
0.0222094548 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 18 |
Hb_002817_020 |
0.0225788505 |
- |
- |
- |
| 19 |
Hb_000769_040 |
0.0237708398 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 20 |
Hb_000221_010 |
0.026780885 |
- |
- |
polyprotein [Oryza australiensis] |