| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
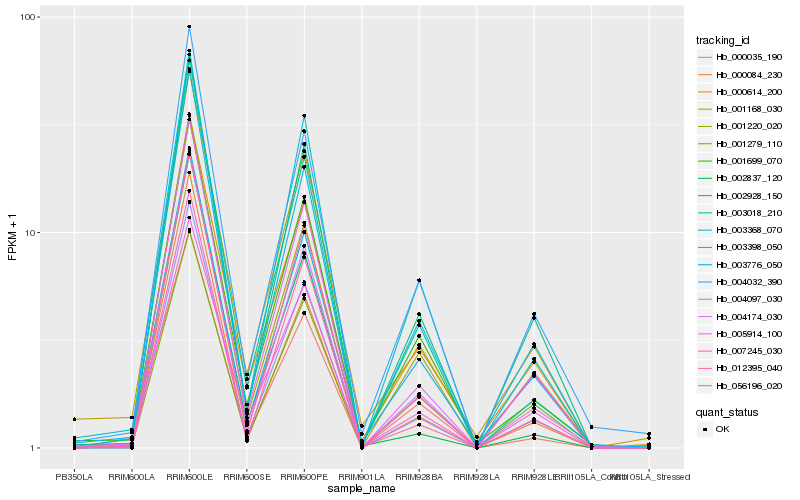
Hb_004097_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 2 |
Hb_005914_100 |
0.0260896144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004174_030 |
0.0372715971 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Populus euphratica] |
| 4 |
Hb_002928_150 |
0.0518043571 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 5 |
Hb_003398_050 |
0.0556991185 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 6 |
Hb_056196_020 |
0.0604016025 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003018_210 |
0.0610772889 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 8 |
Hb_001220_020 |
0.0680535493 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 9 |
Hb_007245_030 |
0.0712571526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004032_390 |
0.0791861043 |
- |
- |
CDK, putative [Ricinus communis] |
| 11 |
Hb_003776_050 |
0.0822563614 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 12 |
Hb_000614_200 |
0.0843447648 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 13 |
Hb_003368_070 |
0.0844170765 |
- |
- |
PREDICTED: uncharacterized protein LOC105109387 [Populus euphratica] |
| 14 |
Hb_001699_070 |
0.0859071742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001168_030 |
0.0879454149 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 16 |
Hb_000035_190 |
0.0919287991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000084_230 |
0.0919800446 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 18 |
Hb_002837_120 |
0.092395027 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 19 |
Hb_012395_040 |
0.0926733127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001279_110 |
0.0931561584 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |