| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
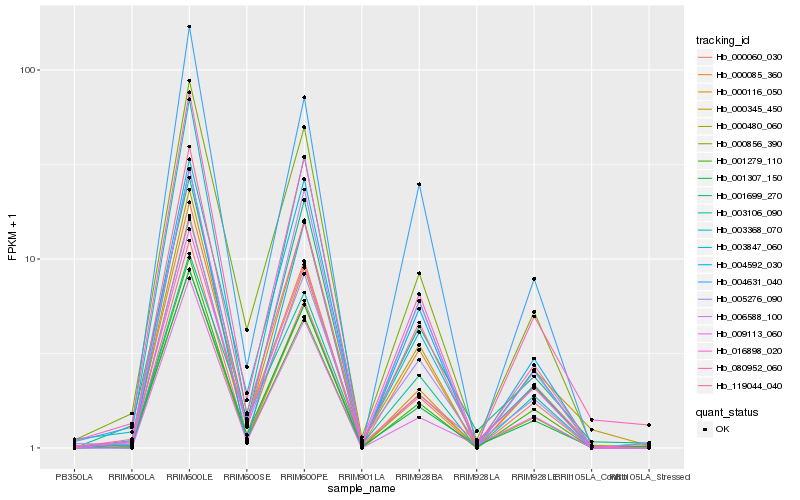
Hb_000480_060 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 2 |
Hb_001307_150 |
0.0522768182 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_009113_060 |
0.0688294948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001699_270 |
0.0699850099 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 5 |
Hb_006588_100 |
0.0701791693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000345_450 |
0.0704256647 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 7 |
Hb_000085_360 |
0.0707743481 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 8 |
Hb_005276_090 |
0.0716927749 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000856_390 |
0.0729134097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000116_050 |
0.0787041437 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_003847_060 |
0.0796611486 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_016898_020 |
0.0799417724 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 13 |
Hb_004592_030 |
0.0804254325 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_001279_110 |
0.0835024852 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 15 |
Hb_004631_040 |
0.0890301583 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 16 |
Hb_080952_060 |
0.0893056691 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 17 |
Hb_000060_030 |
0.089546272 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like [Jatropha curcas] |
| 18 |
Hb_003368_070 |
0.0901101723 |
- |
- |
PREDICTED: uncharacterized protein LOC105109387 [Populus euphratica] |
| 19 |
Hb_003106_090 |
0.0904522553 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 20 |
Hb_119044_040 |
0.0915192492 |
- |
- |
cyclin B, putative [Ricinus communis] |