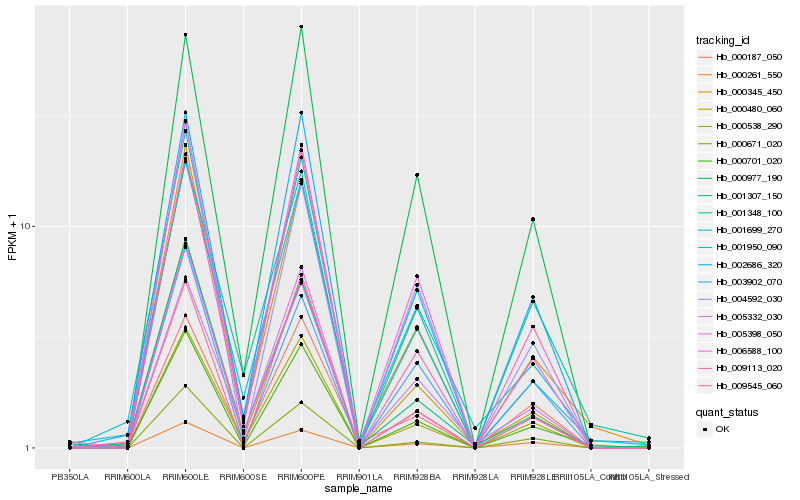
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000701_020 |
0.0 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000345_450 |
0.0711846562 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 3 |
Hb_002686_320 |
0.0795401688 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_000538_290 |
0.0892596932 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 5 |
Hb_006588_100 |
0.0900135671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000977_190 |
0.0945894432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000671_020 |
0.0965020172 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |
| 8 |
Hb_009113_020 |
0.0979493781 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 9 |
Hb_009545_060 |
0.1020159963 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 10 |
Hb_000480_060 |
0.1033936657 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 11 |
Hb_001699_270 |
0.1062770642 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 12 |
Hb_000261_550 |
0.1071456719 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 13 |
Hb_001950_090 |
0.1073919675 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 14 |
Hb_005398_050 |
0.1140998859 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001348_100 |
0.1142120164 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 16 |
Hb_004592_030 |
0.1163957471 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_000187_050 |
0.116622667 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 18 |
Hb_005332_030 |
0.1182812546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003902_070 |
0.1210540235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001307_150 |
0.1221374793 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |