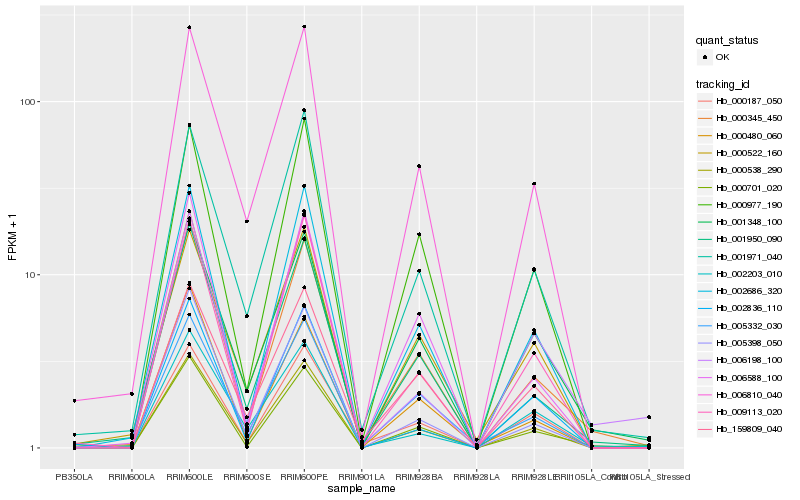
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_290 |
0.0 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 2 |
Hb_002686_320 |
0.0677695014 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 3 |
Hb_009113_020 |
0.0733840141 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 4 |
Hb_006810_040 |
0.08666794 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000701_020 |
0.0892596932 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000977_190 |
0.0915372245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000187_050 |
0.09224411 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 8 |
Hb_005398_050 |
0.0932839512 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001348_100 |
0.0951340645 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 10 |
Hb_001971_040 |
0.095794284 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 11 |
Hb_001950_090 |
0.0981801318 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 12 |
Hb_002836_110 |
0.0984909277 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 13 |
Hb_000345_450 |
0.0998230152 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 14 |
Hb_002203_010 |
0.1039803282 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 15 |
Hb_000480_060 |
0.103985807 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 16 |
Hb_005332_030 |
0.1040784936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_159809_040 |
0.1049540943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006588_100 |
0.1057781357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000522_160 |
0.1060854901 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 20 |
Hb_006198_100 |
0.1067501258 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |