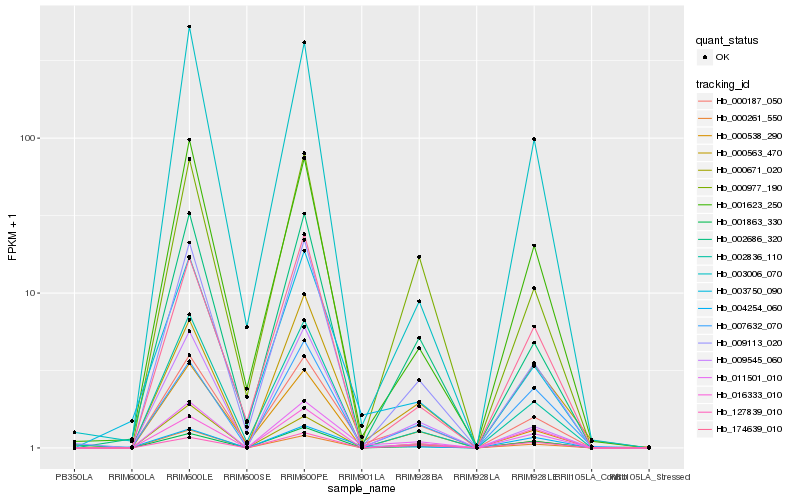
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011501_010 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 2 |
Hb_000187_050 |
0.0879927815 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 3 |
Hb_000563_470 |
0.1231735027 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 4 |
Hb_009113_020 |
0.1300486892 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 5 |
Hb_003750_090 |
0.1314764579 |
- |
- |
BnaC02g34020D [Brassica napus] |
| 6 |
Hb_001863_330 |
0.133049578 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002686_320 |
0.1338470109 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_001623_250 |
0.1353026282 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 9 |
Hb_002836_110 |
0.1373943379 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 10 |
Hb_000261_550 |
0.1389360298 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 11 |
Hb_000671_020 |
0.141310808 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |
| 12 |
Hb_016333_010 |
0.1455335558 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 13 |
Hb_009545_060 |
0.1468561148 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 14 |
Hb_174639_010 |
0.1476463226 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_004254_060 |
0.1482064245 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 16 |
Hb_007632_070 |
0.1486218017 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_000538_290 |
0.1500939109 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 18 |
Hb_127839_010 |
0.1504181572 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 19 |
Hb_003006_070 |
0.1504670503 |
- |
- |
aquaporin [Manihot esculenta] |
| 20 |
Hb_000977_190 |
0.1507204546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |