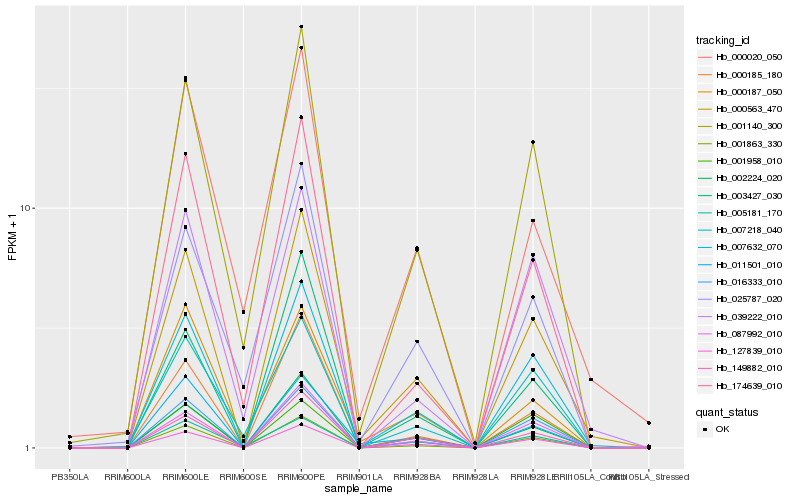
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_470 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 2 |
Hb_007632_070 |
0.0650430884 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_127839_010 |
0.0852735302 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 4 |
Hb_001863_330 |
0.0911704583 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002224_020 |
0.0925013529 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 6 |
Hb_149882_010 |
0.097958544 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 7 |
Hb_016333_010 |
0.0995243266 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 8 |
Hb_087992_010 |
0.1025266645 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_001140_300 |
0.1130908885 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 10 |
Hb_000185_180 |
0.123089925 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 11 |
Hb_011501_010 |
0.1231735027 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 12 |
Hb_003427_030 |
0.1329482659 |
- |
- |
- |
| 13 |
Hb_174639_010 |
0.1358737255 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_005181_170 |
0.1362263347 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 15 |
Hb_039222_010 |
0.1404451864 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000187_050 |
0.1456181394 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 17 |
Hb_025787_020 |
0.1494175008 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_007218_040 |
0.1496138261 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 19 |
Hb_000020_050 |
0.1504347231 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 20 |
Hb_001958_010 |
0.1505865988 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |