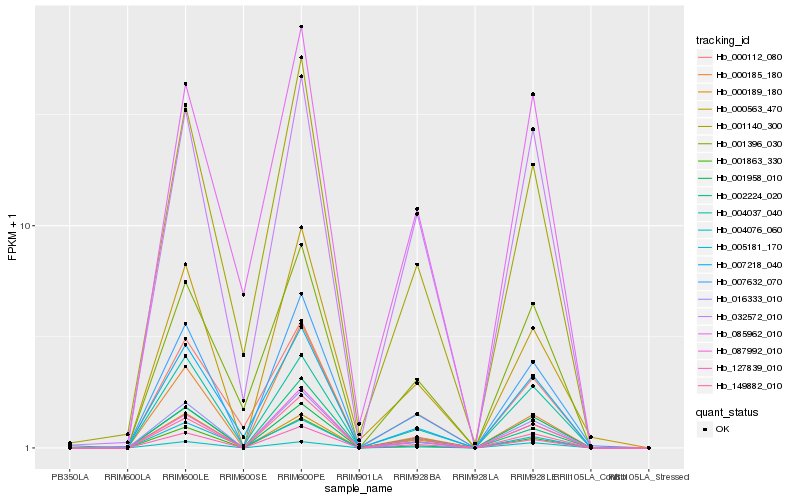
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127839_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 2 |
Hb_016333_010 |
0.0617194443 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 3 |
Hb_007632_070 |
0.0630506627 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_087992_010 |
0.0753589381 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_149882_010 |
0.0776675041 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 6 |
Hb_001863_330 |
0.0794369893 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002224_020 |
0.0841860053 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 8 |
Hb_000563_470 |
0.0852735302 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 9 |
Hb_001958_010 |
0.0952136203 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 10 |
Hb_001140_300 |
0.1000902898 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 11 |
Hb_032572_010 |
0.1025299388 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 12 |
Hb_005181_170 |
0.1056734953 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 13 |
Hb_000189_180 |
0.107139619 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 14 |
Hb_004037_040 |
0.1123595727 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 15 |
Hb_007218_040 |
0.1223710529 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 16 |
Hb_001396_030 |
0.1324810398 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 17 |
Hb_085962_010 |
0.1332103631 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 18 |
Hb_000112_080 |
0.1368467916 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 19 |
Hb_000185_180 |
0.1372303677 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 20 |
Hb_004076_060 |
0.1389633121 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |