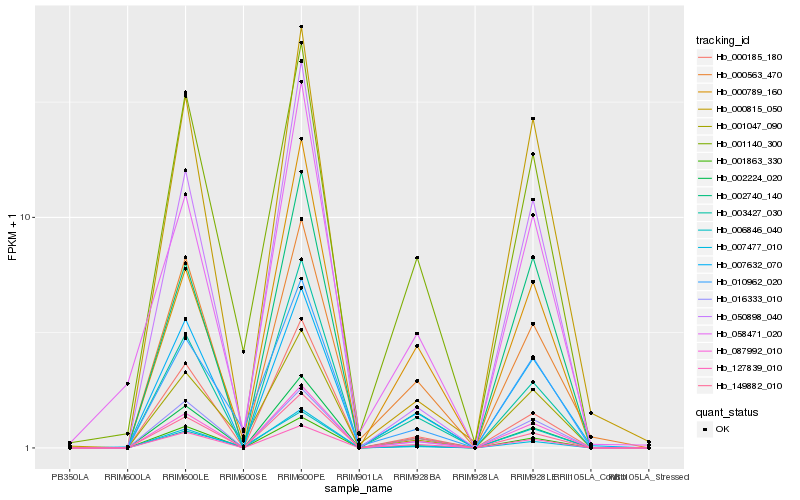
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_087992_010 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_002224_020 |
0.0685376047 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 3 |
Hb_127839_010 |
0.0753589381 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 4 |
Hb_001863_330 |
0.076579937 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 5 |
Hb_016333_010 |
0.0817777159 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 6 |
Hb_007632_070 |
0.0825847618 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_149882_010 |
0.0850854102 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 8 |
Hb_002740_140 |
0.0932438484 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 9 |
Hb_003427_030 |
0.0940359305 |
- |
- |
- |
| 10 |
Hb_000563_470 |
0.1025266645 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 11 |
Hb_001140_300 |
0.1047777194 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 12 |
Hb_000185_180 |
0.1093586953 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 13 |
Hb_001047_090 |
0.1099340296 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 14 |
Hb_010962_020 |
0.1231088632 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 15 |
Hb_000815_050 |
0.1269473982 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 16 |
Hb_007477_010 |
0.1277983223 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 17 |
Hb_000789_160 |
0.1296972484 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 18 |
Hb_050898_040 |
0.1297457349 |
- |
- |
- |
| 19 |
Hb_006846_040 |
0.1302536116 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 20 |
Hb_058471_020 |
0.1315997892 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |