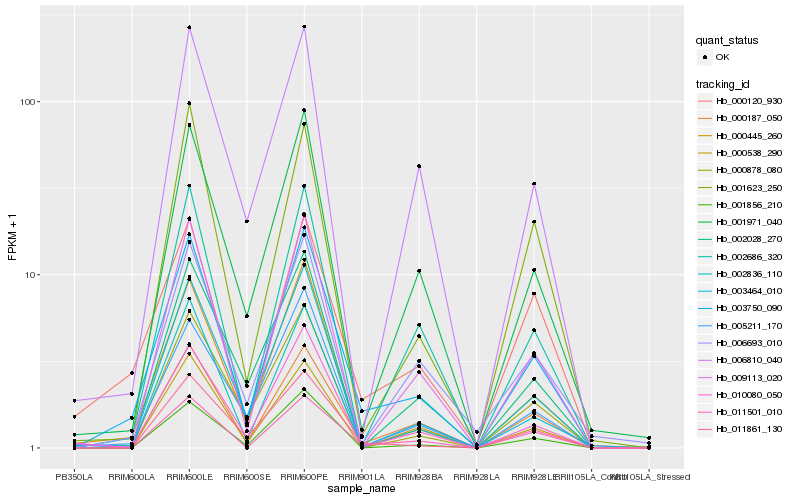
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003750_090 |
0.0 |
- |
- |
BnaC02g34020D [Brassica napus] |
| 2 |
Hb_000187_050 |
0.1172890792 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 3 |
Hb_000445_260 |
0.1191759291 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 4 |
Hb_011861_130 |
0.1238036806 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 5 |
Hb_002686_320 |
0.1281133991 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 6 |
Hb_001971_040 |
0.128356427 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 7 |
Hb_009113_020 |
0.1288807 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 8 |
Hb_010080_050 |
0.1311620353 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 9 |
Hb_011501_010 |
0.1314764579 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 10 |
Hb_001856_210 |
0.1318920435 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 11 |
Hb_002836_110 |
0.1379782105 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 12 |
Hb_003464_010 |
0.1405393455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005211_170 |
0.1408745689 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 14 |
Hb_006693_010 |
0.1419505034 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 15 |
Hb_000120_930 |
0.1436518775 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002028_270 |
0.1441845332 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 17 |
Hb_001623_250 |
0.1447842922 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 18 |
Hb_000538_290 |
0.1457831249 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 19 |
Hb_000878_080 |
0.1460134779 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 20 |
Hb_006810_040 |
0.1475399275 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |