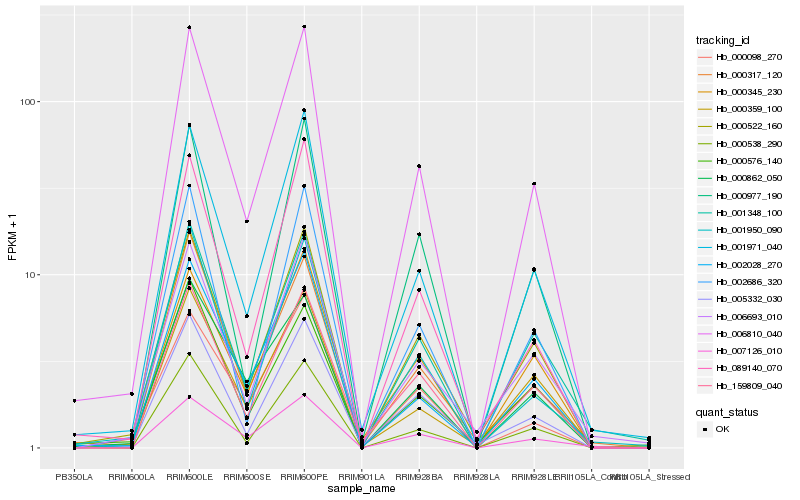
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006810_040 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001971_040 |
0.0610364902 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 3 |
Hb_000522_160 |
0.067598483 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 4 |
Hb_002028_270 |
0.0751701639 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_159809_040 |
0.0790428468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_089140_070 |
0.082346782 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 7 |
Hb_005332_030 |
0.0844781538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000538_290 |
0.08666794 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 9 |
Hb_001950_090 |
0.0906699692 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 10 |
Hb_002686_320 |
0.0914095416 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_000317_120 |
0.093040498 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 12 |
Hb_000345_230 |
0.0951053895 |
- |
- |
- |
| 13 |
Hb_000977_190 |
0.095200927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001348_100 |
0.0956569052 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 15 |
Hb_007126_010 |
0.0985858905 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 16 |
Hb_006693_010 |
0.0986393907 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 17 |
Hb_000359_100 |
0.1009186391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000862_050 |
0.1009510711 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 19 |
Hb_000576_140 |
0.1049551842 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 20 |
Hb_000098_270 |
0.1051279015 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |