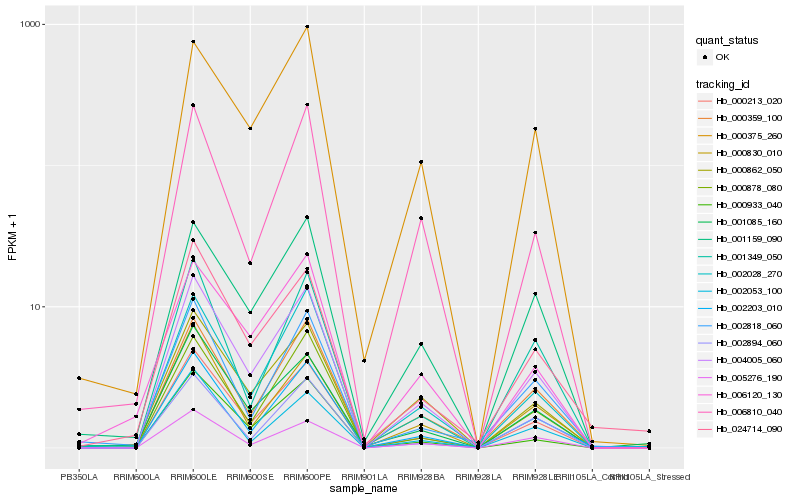
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004005_060 |
0.0 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 2 |
Hb_000213_020 |
0.0588588996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002203_010 |
0.0848687001 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 4 |
Hb_000359_100 |
0.0859221991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002028_270 |
0.0868004426 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000862_050 |
0.0913532918 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 7 |
Hb_000878_080 |
0.0929947475 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 8 |
Hb_000933_040 |
0.0991082613 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 9 |
Hb_001159_090 |
0.0996023957 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 10 |
Hb_005276_190 |
0.1041439213 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000375_260 |
0.1059286579 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 12 |
Hb_001085_160 |
0.1124207483 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 13 |
Hb_000830_010 |
0.1143709303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006810_040 |
0.1170299691 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002894_060 |
0.1172025169 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 16 |
Hb_024714_090 |
0.1178925609 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 17 |
Hb_001349_050 |
0.1196533745 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 18 |
Hb_006120_130 |
0.1196730516 |
- |
- |
- |
| 19 |
Hb_002818_060 |
0.1243805096 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_002053_100 |
0.1249278986 |
- |
- |
actin binding protein, putative [Ricinus communis] |