| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
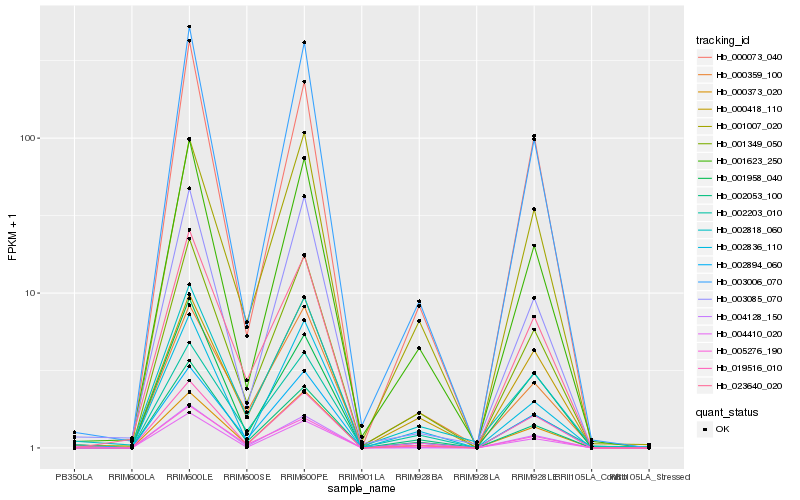
Hb_001349_050 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 2 |
Hb_002894_060 |
0.0467282427 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 3 |
Hb_001623_250 |
0.060606888 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 4 |
Hb_002818_060 |
0.0763246966 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_003006_070 |
0.0783797431 |
- |
- |
aquaporin [Manihot esculenta] |
| 6 |
Hb_019516_010 |
0.0807670513 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 7 |
Hb_002053_100 |
0.081890465 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 8 |
Hb_001007_020 |
0.0820311996 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002203_010 |
0.0841515107 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 10 |
Hb_000418_110 |
0.0844473287 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 11 |
Hb_005276_190 |
0.0870530432 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002836_110 |
0.0891906661 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 13 |
Hb_001958_040 |
0.0941843059 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 14 |
Hb_003085_070 |
0.0962247272 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_004128_150 |
0.0966076743 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 16 |
Hb_000359_100 |
0.1004653812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000373_020 |
0.1004814122 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 18 |
Hb_000073_040 |
0.1015224647 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 19 |
Hb_023640_020 |
0.1030447755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004410_020 |
0.105107187 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |