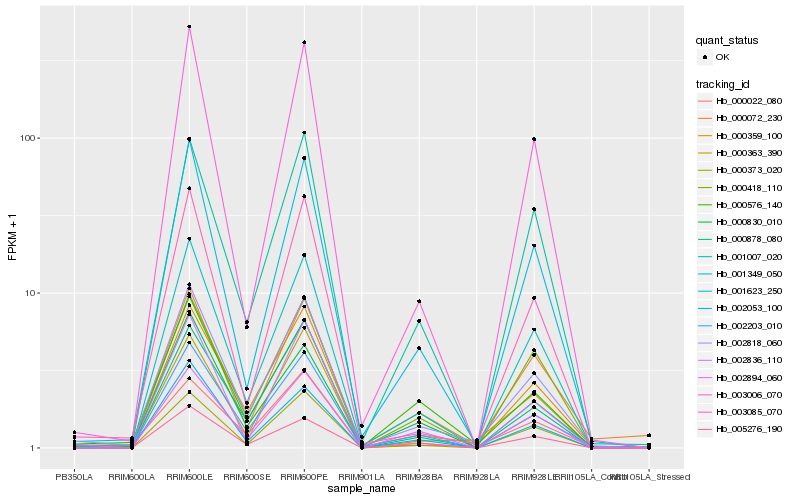
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002818_060 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001349_050 |
0.0763246966 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 3 |
Hb_001623_250 |
0.0897341494 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 4 |
Hb_002203_010 |
0.0910184005 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 5 |
Hb_000359_100 |
0.0915133431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002894_060 |
0.0934361617 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 7 |
Hb_000363_390 |
0.0973624378 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002836_110 |
0.1029825397 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 9 |
Hb_000373_020 |
0.1040505967 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 10 |
Hb_003085_070 |
0.1062101717 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000576_140 |
0.1094993682 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 12 |
Hb_000022_080 |
0.1106683923 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 13 |
Hb_001007_020 |
0.1107794627 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000072_230 |
0.1114130983 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |
| 15 |
Hb_002053_100 |
0.1134274573 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 16 |
Hb_000830_010 |
0.1139975811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000878_080 |
0.1143673971 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 18 |
Hb_003006_070 |
0.1144819851 |
- |
- |
aquaporin [Manihot esculenta] |
| 19 |
Hb_005276_190 |
0.1158939092 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000418_110 |
0.116112951 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |