| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
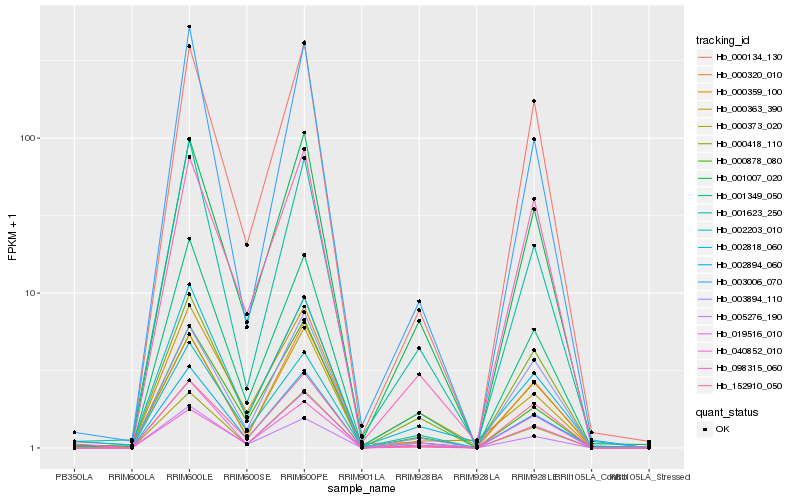
Hb_002894_060 |
0.0 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 2 |
Hb_001349_050 |
0.0467282427 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 3 |
Hb_001007_020 |
0.0550174519 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000418_110 |
0.0583215858 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 5 |
Hb_019516_010 |
0.0759732118 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 6 |
Hb_000134_130 |
0.0780176398 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 7 |
Hb_001623_250 |
0.0896715164 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 8 |
Hb_000878_080 |
0.0907612193 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 9 |
Hb_002203_010 |
0.0927197778 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 10 |
Hb_002818_060 |
0.0934361617 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000373_020 |
0.0959281967 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 12 |
Hb_000359_100 |
0.0967879567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_040852_010 |
0.0977082501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_098315_060 |
0.0980135043 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 15 |
Hb_003006_070 |
0.099126989 |
- |
- |
aquaporin [Manihot esculenta] |
| 16 |
Hb_000320_010 |
0.0994228094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000363_390 |
0.1002441825 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_003894_110 |
0.101469507 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 19 |
Hb_005276_190 |
0.1018045076 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 20 |
Hb_152910_050 |
0.1034691167 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |