| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
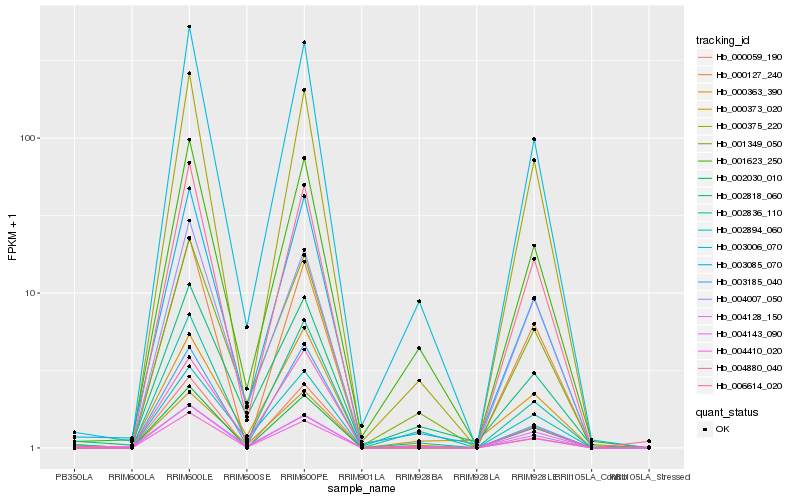
Hb_003085_070 |
0.0 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_003006_070 |
0.0584975222 |
- |
- |
aquaporin [Manihot esculenta] |
| 3 |
Hb_001623_250 |
0.0829779808 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 4 |
Hb_003185_040 |
0.0844275067 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 5 |
Hb_002836_110 |
0.0900035673 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 6 |
Hb_006614_020 |
0.0911609092 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 7 |
Hb_000375_220 |
0.0941397983 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 8 |
Hb_004410_020 |
0.0953354525 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 9 |
Hb_004128_150 |
0.0954758277 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 10 |
Hb_001349_050 |
0.0962247272 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 11 |
Hb_004880_040 |
0.0990210667 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002030_010 |
0.09954976 |
- |
- |
hypothetical protein JCGZ_18681 [Jatropha curcas] |
| 13 |
Hb_002818_060 |
0.1062101717 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000363_390 |
0.1063553989 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002894_060 |
0.1095941634 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 16 |
Hb_000373_020 |
0.1150303321 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 17 |
Hb_000127_240 |
0.1159062411 |
- |
- |
SAUR39 [Populus tomentosa] |
| 18 |
Hb_000059_190 |
0.1167820635 |
- |
- |
PREDICTED: uncharacterized protein LOC105121874 [Populus euphratica] |
| 19 |
Hb_004143_090 |
0.1204456141 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004007_050 |
0.120474392 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |