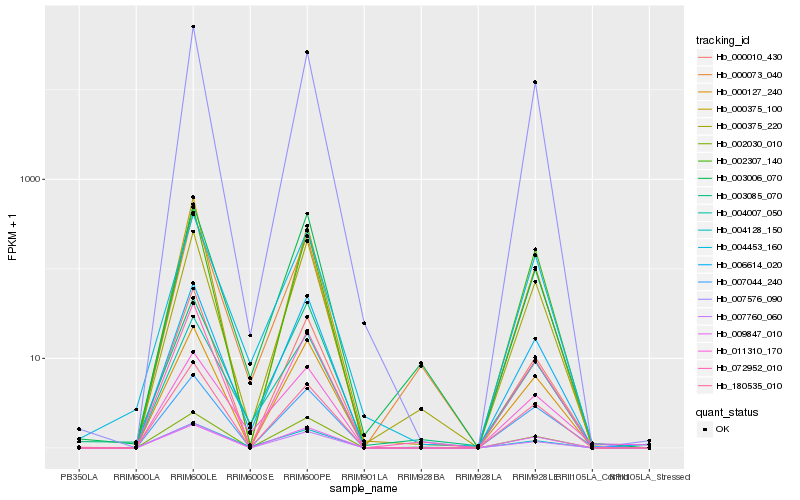
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006614_020 |
0.0 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 2 |
Hb_000127_240 |
0.0500769093 |
- |
- |
SAUR39 [Populus tomentosa] |
| 3 |
Hb_002030_010 |
0.0521498853 |
- |
- |
hypothetical protein JCGZ_18681 [Jatropha curcas] |
| 4 |
Hb_000375_220 |
0.0571274427 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 5 |
Hb_004128_150 |
0.059021314 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 6 |
Hb_007760_060 |
0.0625547277 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 7 |
Hb_004007_050 |
0.0748785845 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 8 |
Hb_003006_070 |
0.0775979239 |
- |
- |
aquaporin [Manihot esculenta] |
| 9 |
Hb_007576_090 |
0.0801285386 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 10 |
Hb_180535_010 |
0.087259915 |
- |
- |
- |
| 11 |
Hb_004453_160 |
0.0894511757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003085_070 |
0.0911609092 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000073_040 |
0.0926341947 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 14 |
Hb_007044_240 |
0.0936995056 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009847_010 |
0.0937517035 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 16 |
Hb_000375_100 |
0.0943904589 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 17 |
Hb_011310_170 |
0.0977778759 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000010_430 |
0.0981408381 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 19 |
Hb_072952_010 |
0.098197862 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 20 |
Hb_002307_140 |
0.098749061 |
transcription factor |
TF Family: AUX/IAA |
aux/IAA family protein [Populus trichocarpa] |