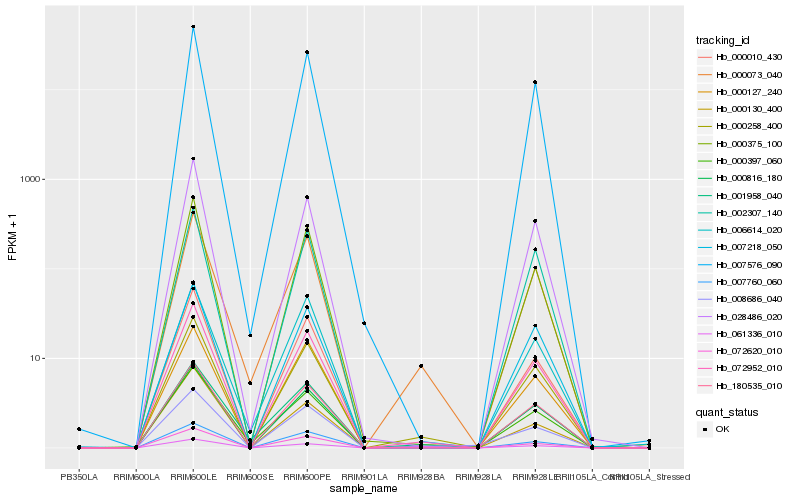
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_072952_010 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 2 |
Hb_061336_010 |
0.0411854806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007576_090 |
0.0439123486 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 4 |
Hb_000375_100 |
0.0470865947 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 5 |
Hb_180535_010 |
0.0475879967 |
- |
- |
- |
| 6 |
Hb_000816_180 |
0.0492699008 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 7 |
Hb_000258_400 |
0.0495245903 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 8 |
Hb_000010_430 |
0.0535833221 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_007760_060 |
0.0552205803 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 10 |
Hb_028486_020 |
0.0594897494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007218_050 |
0.0703617541 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000073_040 |
0.0722260391 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 13 |
Hb_000127_240 |
0.078027094 |
- |
- |
SAUR39 [Populus tomentosa] |
| 14 |
Hb_002307_140 |
0.0794931461 |
transcription factor |
TF Family: AUX/IAA |
aux/IAA family protein [Populus trichocarpa] |
| 15 |
Hb_000397_060 |
0.0798884427 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 16 |
Hb_072620_010 |
0.0847143253 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 17 |
Hb_000130_400 |
0.0880924769 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0001s38320g [Populus trichocarpa] |
| 18 |
Hb_008686_040 |
0.0924224655 |
- |
- |
PREDICTED: uncharacterized protein LOC105632278 isoform X3 [Jatropha curcas] |
| 19 |
Hb_001958_040 |
0.0941989983 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 20 |
Hb_006614_020 |
0.098197862 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |