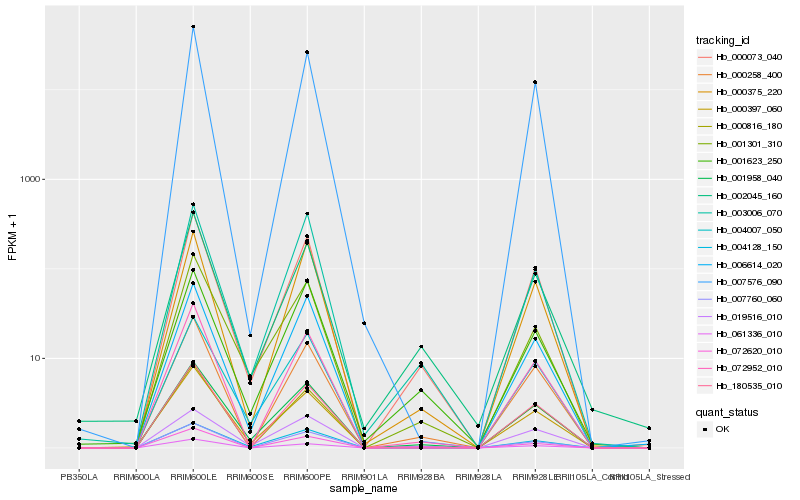
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000073_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 2 |
Hb_001958_040 |
0.044333587 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 3 |
Hb_000397_060 |
0.0609905883 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 4 |
Hb_000258_400 |
0.0662456296 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 5 |
Hb_072952_010 |
0.0722260391 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 6 |
Hb_072620_010 |
0.0767020275 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 7 |
Hb_002045_160 |
0.0781667179 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 8 |
Hb_004007_050 |
0.0827386739 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 9 |
Hb_019516_010 |
0.0831034815 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000375_220 |
0.0850009317 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 11 |
Hb_007576_090 |
0.0851544546 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 12 |
Hb_180535_010 |
0.0885945955 |
- |
- |
- |
| 13 |
Hb_001623_250 |
0.0916111419 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 14 |
Hb_001301_310 |
0.0916564028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006614_020 |
0.0926341947 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 16 |
Hb_000816_180 |
0.0931120154 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 17 |
Hb_007760_060 |
0.0936995382 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 18 |
Hb_003006_070 |
0.093789355 |
- |
- |
aquaporin [Manihot esculenta] |
| 19 |
Hb_061336_010 |
0.094489965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004128_150 |
0.0952823427 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |