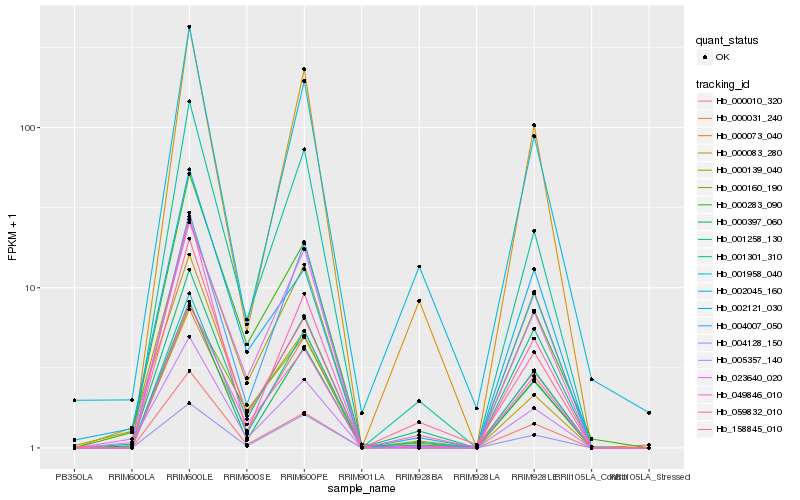
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005357_140 |
0.0 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 2 |
Hb_000397_060 |
0.071496313 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 3 |
Hb_000139_040 |
0.0884416773 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 4 |
Hb_001301_310 |
0.093332548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_049846_010 |
0.0974913561 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 6 |
Hb_001958_040 |
0.0978565341 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 7 |
Hb_000283_090 |
0.1016341233 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_023640_020 |
0.104478021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000010_320 |
0.1046564021 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 10 |
Hb_004007_050 |
0.1159881327 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 11 |
Hb_000073_040 |
0.1191569011 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 12 |
Hb_059832_010 |
0.1199956069 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 13 |
Hb_000083_280 |
0.120796637 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_002045_160 |
0.1214900801 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 15 |
Hb_000031_240 |
0.1235811063 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 16 |
Hb_001258_130 |
0.1244131489 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 17 |
Hb_158845_010 |
0.1257957358 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 18 |
Hb_004128_150 |
0.1258605167 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 19 |
Hb_002121_030 |
0.1271219528 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 20 |
Hb_000160_190 |
0.1281917836 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |