Hb_004003_020
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig4003: 19854-28755 |
| Sequence |   |
Annotation
kegg
| ID | egr:104441780 |
|---|---|
| description | protein NP24-like |
nr
| ID | CDP16244.1 |
|---|---|
| description | unnamed protein product [Coffea canephora] |
swissprot
| ID | P81370 |
|---|---|
| description | Thaumatin-like protein OS=Actinidia deliciosa GN=tlp PE=1 SV=2 |
trembl
| ID | A0A068V707 |
|---|---|
| description | Coffea canephora DH200=94 genomic scaffold, scaffold_120 OS=Coffea canephora GN=GSCOC_T00018006001 PE=4 SV=1 |
Gene Ontology
| ID | - |
|---|---|
| description | - |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_39814: 22991-24012 |
|---|---|
| cDNA (Sanger) (ID:Location) |
003_G24.ab1: 23221-24001 , 003_L07.ab1: 23260-24010 , 004_K23.ab1: 23218-24010 , 004_L24.ab1: 23202-24001 , 007_D20.ab1: 23205-24010 , 013_J07.ab1: 23224-24010 , 016_B20.ab1: 23225-24001 , 016_K18.ab1: 23287-24008 , 019_G09.ab1: 23401-24010 , 021_D13.ab1: 23224-24008 , 021_M02.ab1: 23224-24012 , 025_K18.ab1: 23230-24010 , 033_L12.ab1: 23257-24012 , 034_D22.ab1: 23208-24006 , 034_E01.ab1: 23226-24010 , 036_A02.ab1: 23223-24016 , 036_B17.ab1: 23247-24008 , 036_L13.ab1: 23211-23987 , 038_L01.ab1: 23223-24010 , 042_C01.ab1: 23209-24008 , 042_H02.ab1: 23225-24008 , 047_C02.ab1: 23225-24008 , 051_A06.ab1: 23300-24010 |
Similar expressed genes (Top20)
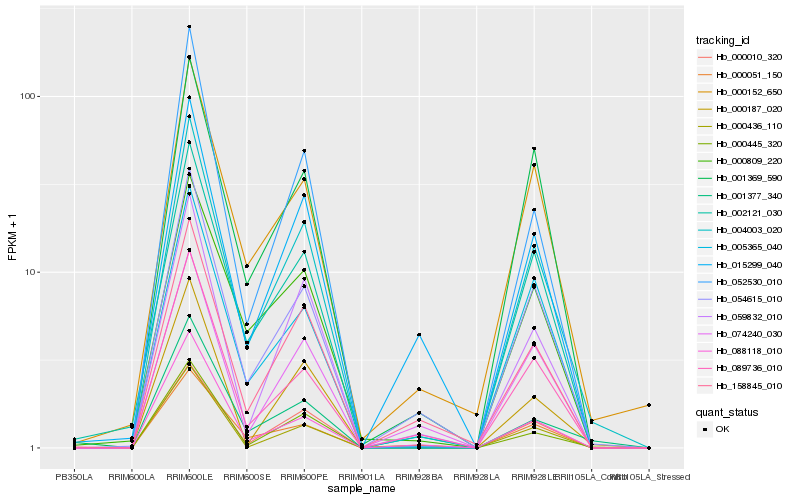
Gene co-expression network
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 0 | 2.75317 | 76.0148 | 18.3063 | 0 | 0 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 0.405829 | 0 | 0 | 0.16118 | 13.1564 |

