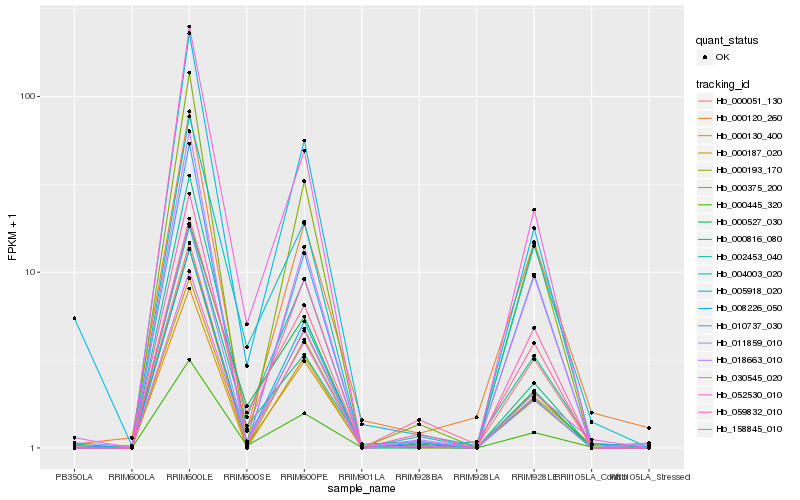
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_320 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 2 |
Hb_000187_020 |
0.0550070779 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_002453_040 |
0.073427276 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 4 |
Hb_052530_010 |
0.0795163616 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 5 |
Hb_059832_010 |
0.0799748591 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 6 |
Hb_000193_170 |
0.0840222811 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 7 |
Hb_005918_020 |
0.0862331846 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 8 |
Hb_008226_050 |
0.086280341 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 9 |
Hb_000375_200 |
0.0903370045 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 10 |
Hb_030545_020 |
0.0903787191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004003_020 |
0.0917175337 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000051_130 |
0.0941198012 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 13 |
Hb_000527_030 |
0.0975284683 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 14 |
Hb_000130_400 |
0.1005022406 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0001s38320g [Populus trichocarpa] |
| 15 |
Hb_011859_010 |
0.1066384363 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 16 |
Hb_000120_260 |
0.1095299928 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 17 |
Hb_018663_010 |
0.1097000956 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 18 |
Hb_000816_080 |
0.1099853146 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 19 |
Hb_010737_030 |
0.1101333062 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_158845_010 |
0.1118649708 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |